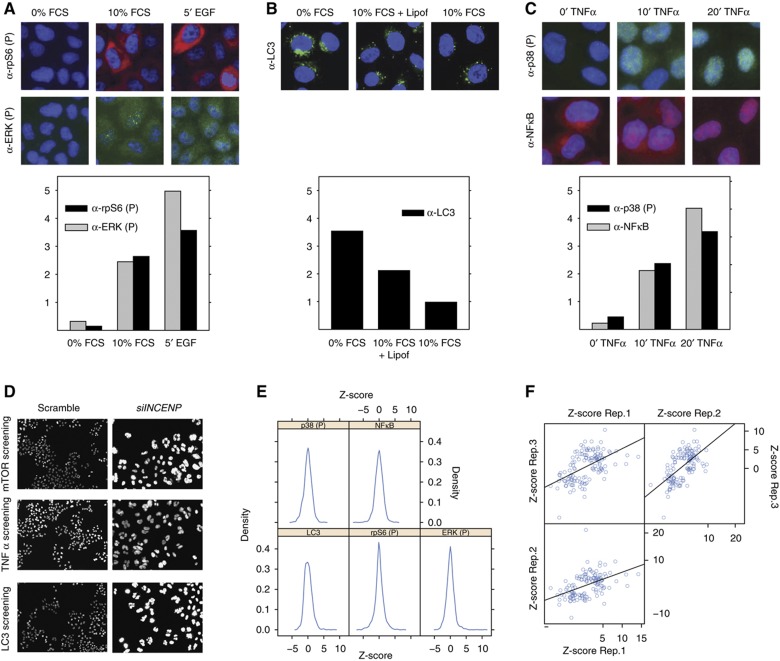
Figure 2.
Identification of phosphatase hits. Preliminary experiments were carried out to identify experimental conditions yielding intermediate readout values. (A) HeLa cells were starved for 6 h (0% FCS), induced with EGF for 5 min (5′ EGF) or left untreated (10% FCS). Cells were fixed and stained with anti-phospho ERK (FITC), anti-phospho rpS6 (TRITC) antibodies and DAPI to visualize nuclei. Cells were analyzed by indirect fluorescence microscopy coupled to automated image analysis. The ERK phosphorylation level in the nucleus (gray bar) as well as the rpS6 phosphorylation in the cytosol (black bar) were automatically measured and plotted. (B) HeLa cells were serum and amino acids deprived for 3 h (0% FCS) or left untreated (10% FCS) or transfected with an empty vector. Cells were fixed and stained with anti-LC3 antibody (FITC) and DAPI to visualize nuclei. To estimate autophagosome formation, images were analyzed by Cell Profiler and the mean LC3 intensity in the cytosol was measured and plotted as a bar graph. (C) HeLa cells were incubated with TNFα and sampled at time 0, 10 and 20 min. Cells were fixed, stained with anti-phospho p38 and anti-NFκB and visualized by indirect fluorescence microscopy. The nuclear p38 phosphorylation level (black bar) as well as the nuclear translocation of NFκB (gray bar) were measured by the Cell Profiler software and plotted. (D) HeLa cells grown on spots of siRNAs targeting the INCENP gene or a scrambled control were stained with DAPI. Images were acquired by automatic fluorescence microscopy with a × 20 objective. siRNA inhibition of the INCENP gene, causing a clear mitotic phenotype, was used as control of siRNA transfection efficiency (Neumann et al, 2010). (E) Distribution of the Z-scores of the phenotypic readouts of cells silenced in the different phosphatase genes. (F) The Z-score of each pair of three different biological replicates of phosphatase hits was plotted on the X and Y axis of a dispersion plot, respectively.