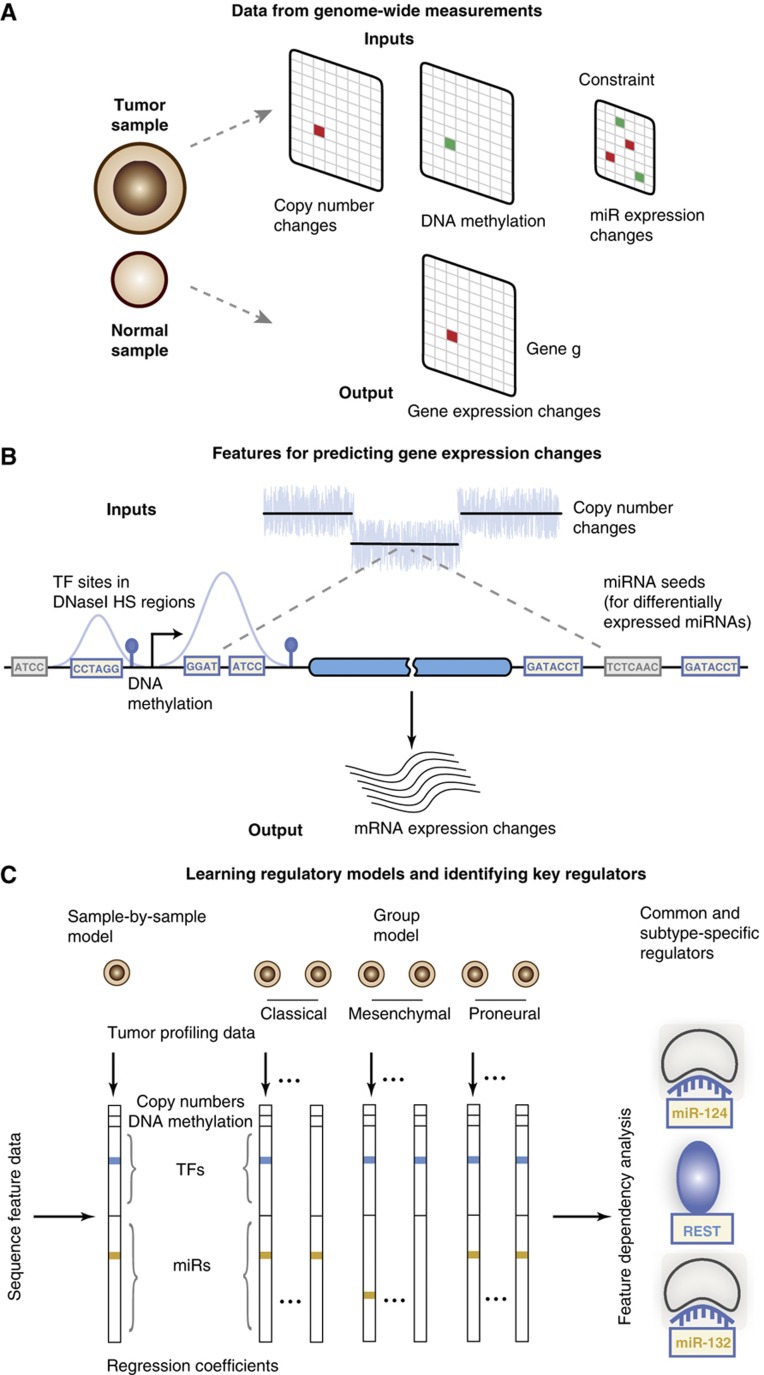
Figure 1.
Modeling gene expression changes in tumors to identify dysregulated transcription factors and microRNAs. (A) Genome-wide measurements like copy number, DNA methylation, and miRNA expression are used to predict gene expression changes of tumor samples relative to normal references. (B) To infer dysregulated regulatory programs from tumor profiling data, change in gene expression in a tumor sample is modeled as linear function of the gene’s copy number, DNA methylation at the promoter (when available for the sample), and counts of transcription factor binding sites in the DNaseI hypersensitive regions of the gene’s promoter and conserved miRNA binding sites in the 3′UTR. (C) The linear model is trained for all tumors, either on a sample-by-sample basis or simultaneously by using a group approach, on all Refseq genes using sparse regression so that only a few explanatory variables have non-zero regression coefficients. In particular, only a small number of transcription factors (TFs) and miRNAs, that is, those whose binding sites best correlate with target gene expression changes in the tumor sample, enter into the regression model. Feature dependency analysis on these regression models identifies common and subtype-specific regulators.