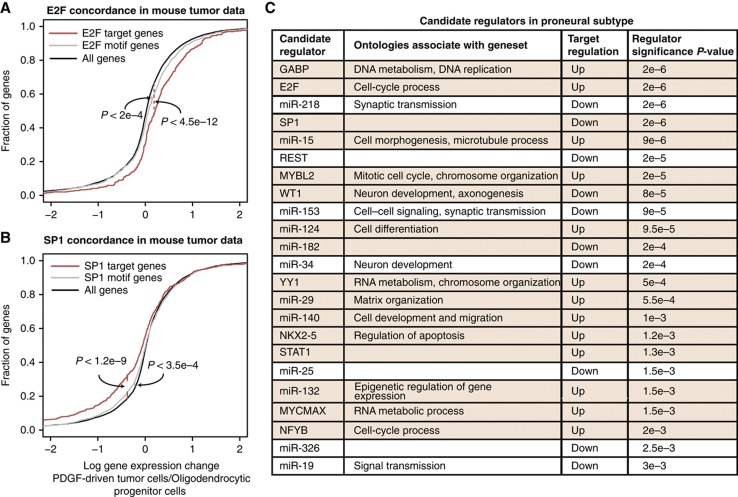
Figure 4.
Gene sets for candidate proneural regulators display coherent functional annotations and consistent in vivo expression changes in PDGF-driven mouse tumors. (A) Targets of E2F, a proneural candidate regulator, show significant upregulation in PDGF-driven Olig2+ mouse tumor cells relative to mouse oligodendrocyte progenitor cells (OPCs) (P<2e−4, Kolmogorov–Smirnov test). Upregulation of the proneural E2F gene set is stronger than the motif-based target set (P<4.5e−12). Human genes were mapped to mouse genes using Homologene. (B) Targets of SP1 show significant downregulation in mouse tumor cells relative to OPCs (P<3.5e−4). Downregulation of proneural SP1 gene set is stronger than motif-based target set (P<1.2e−9). (C) The table lists candidate proneural regulators selected at 10% FDR. Functional annotations were determined by looking for overrepresented terms from the Gene Ontology ‘Biological Process’ in gene sets associated with the candidate regulator. Regulators concordant with PDGF-driven Olig2+ mouse tumor data are shown with rows highlighted in brown. Proneural regulators are ranked by their significance in the regression model, assessed by empirical P-values relative to the previously described randomized models and corrected for multiple-hypothesis testing using the Benjamini-Hochberg procedure.