Abstract
Leaves have a central role in plant energy capture and carbon conversion and therefore must continuously adapt their development to prevailing environmental conditions. To reveal the dynamic systems behaviour of leaf development, we profiled Arabidopsis leaf number six in depth at four different growth stages, at both the end-of-day and end-of-night, in plants growing in two controlled experimental conditions: short-day conditions with optimal soil water content and constant reduced soil water conditions. We found that the lower soil water potential led to reduced, but prolonged, growth and an adaptation at the molecular level without a drought stress response. Clustering of the protein and transcript data using a decision tree revealed different patterns in abundance changes across the growth stages and between end-of-day and end-of-night that are linked to specific biological functions. Correlations between protein and transcript levels depend on the time-of-day and also on protein localisation and function. Surprisingly, only very few of >1700 quantified proteins showed diurnal abundance fluctuations, despite strong fluctuations at the transcript level.
Keywords: adaptation, integrated data analysis, leaf growth, molecular profiling, water deficit
Introduction
Leaves are key organs for plant biomass and seed production because of their roles in energy capture and carbon conversion. Recently, concerns about climate change have raised the awareness of plants being exposed to increasing temperatures and water scarcity (Fedoroff et al, 2010). Signal transduction pathways and transcription factors have been identified that are activated in response to drought and other abiotic stresses (Sakuma et al, 2006). However, a systems-level analysis of growth processes is needed to understand the regulatory network that underlies adaptation to changes in soil water content. Leaves provide the entry point for dissecting adaptive regulatory processes because they must adjust their growth and physiological responses to soil water availability. The dynamics of leaf development and its subsequent growth to a fully mature organ have been studied in different species at both organ and cellular scales, but the underlying molecular mechanisms are not yet fully understood. Typically, a leaf is initiated at the shoot apical meristem at a site of maximum auxin activity (Reinhardt et al, 2003), then the leaf primordium grows through cell division. Subsequent leaf size and shape result from differential patterns of cell proliferation and cell expansion. During the proliferation phase, cells are multiplied through mitotic division and growth is mediated by the increase in cytoplasmic volume accompanied by rapid protein synthesis and active metabolism (Ingram and Waites, 2006). In dicotyledonous plants, cell divisions in the leaf lamella cease progressively, following a spatial gradient from the tip of the leaf to its base that is more or less pronounced depending on the species (Granier and Tardieu, 1998; Donnelly et al, 1999). This cell cycle arrest front moving through the developing leaf is followed by a large increase in cell expansion rate. Cell proliferation continues longer in dispersed stomate-forming meristemoids and in vascular procambium (Donnelly et al, 1999). Cell expansion is driven by uptake of water into the vacuole and controlled modifications and enlargement of the cell wall, and accounts for most of the leaf mass increase in growing plants. It is associated with DNA endoreplication and differentiation into specialised cells (Ingram and Waites, 2006; Anastasiou and Lenhard, 2007; Gonzalez et al, 2012). The impact of environmental constraints, such as light or soil water deficit, on these processes is also well known at the leaf and cell levels in both 2D (Cookson et al, 2006) and 3D (Wuyts et al, 2012).
The successive steps of leaf growth are characterised by distinct spatial and temporal molecular profiles. Specific gene expression patterns during leaf initiation reflect the establishment of polarity and early differentiation events (Beemster et al, 2005; Fleming, 2005; Barkoulas et al, 2007; Hay and Tsiantis, 2009). Recently, transcription data became available for expanding and mature leaves for Arabidopsis leaf 3 (Skirycz et al, 2010) and leaf 7 (Breeze et al, 2011). However, the mechanisms that link gene expression patterns to protein accumulation and regulatory networks are essentially unknown. Large-scale protein profiling data for leaf development have so far not been reported, although differential protein expression and activity are the main determinants of cellular states. Recent studies in various organisms have revealed that protein abundance is regulated at many different levels and that RNA expression dynamics does not necessarily mediate proportional protein abundance changes (de Sousa Abreu et al, 2009; Maier et al, 2009; Piques et al, 2009; Vogel et al, 2010; Lee et al, 2011; Maier et al, 2011; Schwanhäusser et al, 2011). Here, we report an integrated analysis of quantitative transcript and protein measurements at different stages of Arabidopsis leaf development using leaf number 6 to establish how dynamic RNA and protein patterns relate to the phenotypical changes during leaf development. The light–dark cycle leads to recurring fluctuations in the light regime and the supply of carbon (Smith and Stitt, 2007; Usadel et al, 2008) and large changes in leaf expansion rates (Pantin et al, 2011). We have integrated dynamic changes in transcript and protein abundances at the end of the day (EOD) and end of the night (EON) to gain insights into how rapid and recurring changes in environmental conditions modify responses during the developmental program. We have also compared fully integrated data sets for leaf number 6 of plants growing under optimal and reduced soil water conditions to understand how a long-term constant moderate water deficit influences physiological processes and systems-level functions during leaf growth and development.
Results
Scope of the study
The size and shape of individual leaves vary during plant development, with morphological and physiological changes marking the transition between different phases of plant growth (Telfer et al, 1997). To avoid confusing the consequences of the chronology of leaf production with the changes associated with the development of individual leaves, we limited our analyses to leaf 6, which is the first adult leaf of the Arabidopsis thaliana (Col-4) rosette in the short-day condition adopted for this study (8 h of constant illumination, 16 h of darkness). Our primary objective was to quantitatively track and compare the molecular components during growth of a single leaf. Therefore, leaf 6 was harvested at four successive stages of development for the analysis of their transcript and protein profiles. We also investigated how the growth profiles varied during the course of the day by comparing samples collected at the EON and at the EOD, at each developmental stage. We also compared how plants grown under a mild water deficit (SWD) differ from the population maintained in optimal watering conditions (SOW). The SWD conditions applied here subjected the plants to 40% reduced soil water content from early stages of development on and well before harvesting of the earliest stage leaves.
The experimental design addressed multiple challenges. To ensure proper statistical analysis and unless otherwise specified, proteome and transcriptome profiling data were obtained from the same biological samples that were harvested in three independent biological experiments (i.e., three independent replicates). Profiling data were acquired with the AGRONOMICS1 tiling array (Rehrauer et al, 2010) for nuclear-encoded transcription, RT–qPCR for plastid gene transcription, and iTRAQ technology (Ross et al, 2004; Pierce et al, 2008) for quantitative proteomics (see Materials and methods and Supplementary Information). Thousands of plants were necessary in each experiment to provide enough biological material for each time point between leaf emergence and growth completion. To limit spatial and temporal microenvironment heterogeneities, plants were grown in the automated phenotyping platform PHENOPSIS (Granier et al, 2006; Fabre et al, 2011). All phenotypical and molecular profiling data and metadata were integrated within a MySQL relational database and a web site was established for data sharing within the project and for dissemination to the community http://www.agronomics.ethz.ch/.
Reducing soil water content strongly influences leaf growth
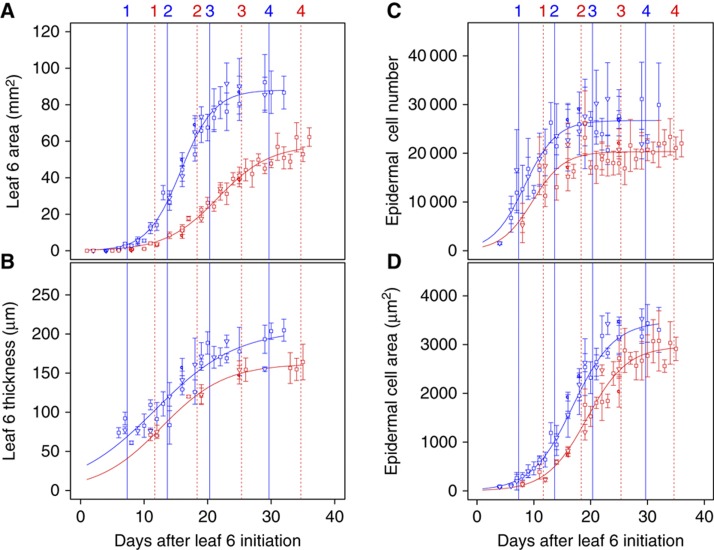
Kinetics of leaf area and thickness expansion were very similar between the three independent replicate experiments for both SOW and SWD conditions, confirming that growth conditions in the PHENOPSIS platform are accurately controlled and results are reproducible across independent successive experiments (Figure 1). A unique sigmoid curve was fitted to the temporal increase in leaf area from leaf initiation until growth cessation that occurred over a period of 28 days in the SOW condition (Figure 1A). Relative area expansion rate was high during the first 10 days following leaf initiation and declined afterwards until expansion ceased. The absolute area expansion rate followed a bell-shaped curve and was highest around 15 days after leaf initiation (Supplementary Table 1). Leaf growth was not synchronous in adaxial–abaxial (blade thickness) and proximal–distal (blade area) dimensions (Figure 1A and B). Rapid adaxial–abaxial growth started very early during development and the leaf already reached one-third of its final thickness when it emerged 7 days after initiation. The absolute thickness expansion rate continued to increase rapidly until 20 days after leaf initiation and thickness reached its maximum a few days after the end of leaf area expansion (Figure 1A; Supplementary Table 1). Based on these profiles, four growth stages were selected for molecular profiling: stage 1, with maximum relative area and thickness expansion rates coinciding with leaf emergence; stage 2, maximum area and thickness absolute expansion rates; stage 3, decreasing leaf area and thickness expansion rates, and stage 4, end of leaf area and thickness expansions.
Figure 1.
Growth phenotypes of leaves harvested for profiling. Kinematic expansion phenotypes of leaves in the SOW (blue) and SWD (red) experiments. Each symbol represents an independent experiment. Leaf 6 changed over time in area (A), thickness (B), epidermal cell number (C) and epidermal cell area (D). Data are presented as mean and s.d. values, n=5. The numbers at the top of the graphs indicate the four growth stages. Source data is available for this figure in the Supplementary Information.
The SWD condition had a marked effect on the area growth and resulted in a 34% reduction in leaf area, whereas final thickness showed a small non-significant reduction of 19% as assessed with a Kruskall–Wallis rank sum test. These decreases were mostly due to reduced area and thickness expansion rates, which were partly offset by an extended growth period (Figure 1A). Because leaf growth was slower in SWD than SOW conditions, the four key stages defined above were delayed accordingly to fit the same dynamics criteria (Figure 1).
Cell number in the adaxial leaf epidermis increased rapidly soon after leaf initiation. At stage 1, cells were dividing actively with little expansion (Figure 1C). Epidermal cell division then decreased, and cell expansion was maximal at stages 2 and 3 (Figure 1C and D; Supplementary Table 1), correlating well with the leaf area increase during these phases. In the SWD condition, the rate but not the duration of epidermal cell division was reduced, resulting in 24% fewer cells compared with the SOW leaf. Additionally, epidermal cell expansion was also slower and final cell size was reduced by 15% in the SWD leaf. Together, these data explain why the extended growth period could not compensate for the reduced final area of the SWD leaf.
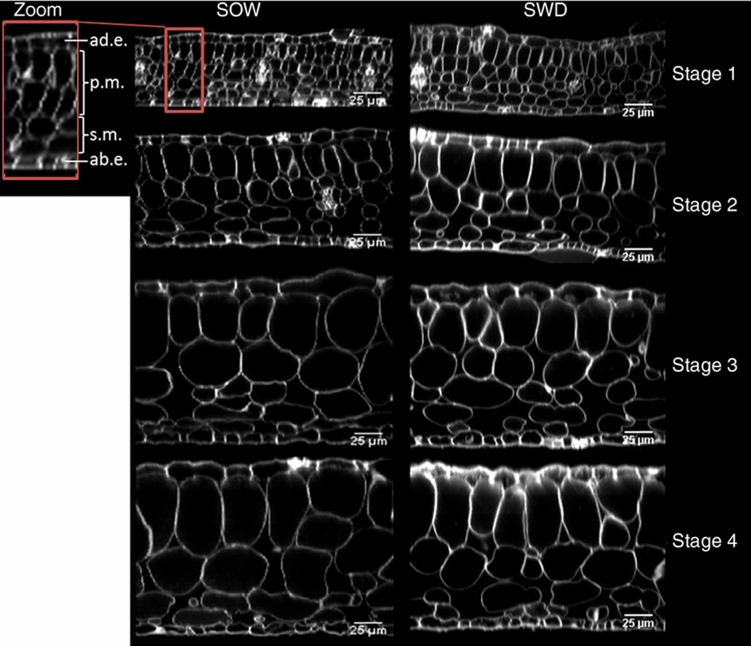
The number of cell layers in the different tissues that contribute primarily to leaf thickness was established early in leaf development before stage 1 and was not affected in the SWD leaf (Figure 2). As expected, the cell density in each tissue was highest at stage 1 and decreased during the subsequent stages (Supplementary Table 2). Consequently, leaf growth in the adaxial–abaxial dimension was mostly the result of cell expansion. Cell densities did not differ between the SOW and SWD leaf for the adaxial and abaxial epidermis and the palisade mesophyll, but cell density in the spongy mesophyll was significantly higher in the SWD leaf. These differences were visible from stage 2 onwards (Figure 2; Supplementary Table 2).
Figure 2.
Transversal sections of leaves across development in SOW and SWD. Sections of leaf 6 in SOW (left panels) and SWD (right panels) were imaged with biphotonic microscopy at the four stages. Tissue layers are marked in the left side zoom section: ad.e.=adaxial epidermis, p.m.=palisade mesophyll, s.m.=spongy mesophyll, ab.e.=abaxial epidermis. Scale bars indicate 25 μm.
DNA ploidy increases during leaf growth but remains lower in leaves growing in water deficit
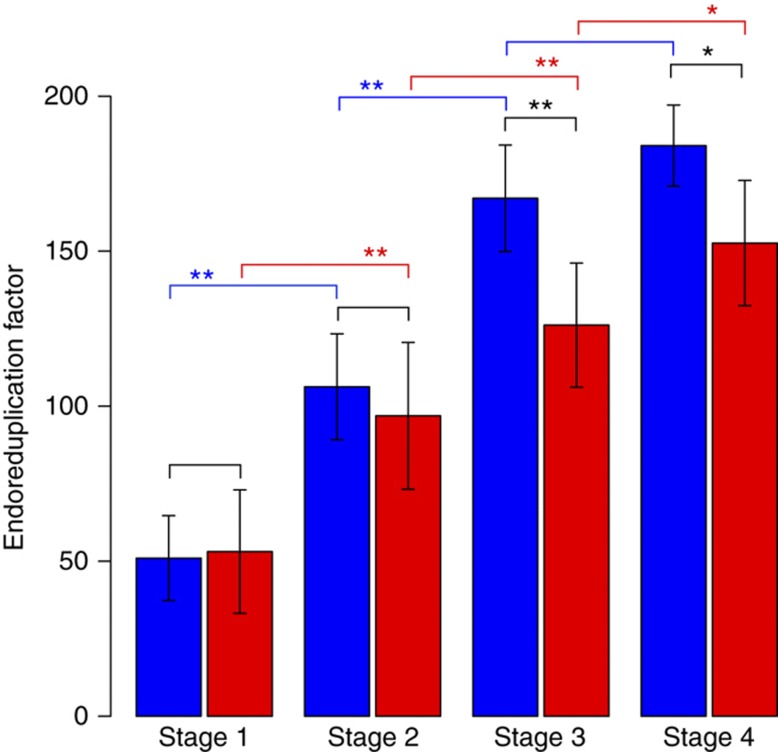
DNA ploidy increased during leaf growth in SOW and SWD leaves, as shown by the increase in the number of endocycles per 100 cells (Figure 3). At stage 1, most cells had a 2C DNA content, but a large proportion had a 4C DNA content and a small number of cells had already gone into two or three successive rounds of endoreduplication. During subsequent stages, the DNA ploidy level then continuously increased (Supplementary Figure 1). In the SWD leaf, the endoreduplication factor was significantly lower later in development compared with the SOW leaf.
Figure 3.
Endoreduplication during leaf development. Endoreduplication factors were calculated and compared for the four development stages in SOW (blue) and SWD (red). Graphs show mean and s.d. values, n≥5; ** indicates statistical significance at level P<0.01, and * at level P<0.05 (two-sided Welch test). Source data is available for this figure in the Supplementary Information.
Dynamics of the leaf transcriptome and the measured proteome vary with growth stage but only the transcriptome varies with time-of-day
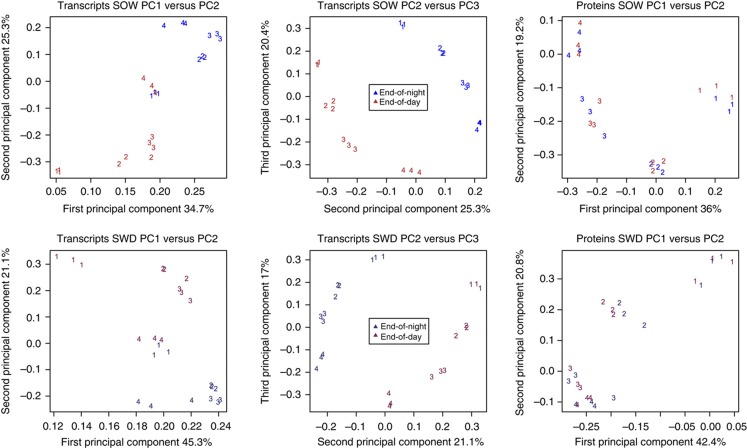
Transcripts of 25 065 genes, of which 22 868 encode proteins, were quantified in SOW leaf samples. The sources of variation measured in the samples were estimated with principal component analysis (PCA) (Figure 4). Notably, the transcriptome PCA first separated the EOD and EON samples (PC1 versus PC2), then the growth stages (PC2 versus PC3), indicating that transcript signatures strongly discriminate both time-of-day and developmental growth stage. The three first components of the PCA explained 35, 25 and 20% of the total variance, respectively. A total of 2081 proteins were quantified via iTRAQ analysis in the same samples. This represents a remarkably large fraction of the proteome that is measurable in a plant organ, especially considering that the leaf has a high dynamic range of protein concentrations (Bindschedler and Cramer, 2011). Based on proteome data, the four growth stages were well separated with the first two principal components, with stage 1 being markedly different (Figure 4). In contrast to the transcripts, the proteome of EOD and EON samples were not separated. Technical variation contributed little to the variance, suggesting that the measurable proteome does not discriminate the time-of-day. Similarly, transcripts of 27 707 genes, of which 24 819 encode proteins, and 1509 proteins were quantified in the SWD leaf samples. In SWD conditions, the transcriptome showed similar PCA patterns to those in SOW conditions, with time-of-day differences accounting for most of the variation, followed by developmental stages (Figure 4). The proteome PCA discriminated growth stages more gradually in SWD compared with SOW samples, and SWD stages 3 and 4 were not separated. The difference in the proteome PCA patterns between SOW and SWD samples can at least in part be explained by the different growth characteristics of SOW and SWD leaves (Figure 1). Together, the PCA suggests similar strong transcription dynamics in SOW and SWD leaves at both time-of-day and different growth stages, which are reflected at the level of the measured proteome regarding growth stages, but not time-of-day.
Figure 4.
PCA of transcript and protein profiles. Upper and lower panels show SOW (upper panels) and SWD (lower panels) experiments, respectively.
Clustering of regulated transcripts and proteins reveals unique patterns during leaf growth
To determine which transcripts and proteins were regulated during leaf development and time-of-day in SOW and SWD leaves, we calculated P-values to assess which transcripts or proteins change in abundance (pGlobal) with a cutoff at pGlobal<0.05. In addition, we calculated the maximum fold-change between the means of each time point and required a fold-change >1.5. These two criteria together minimised the false discovery rate (FDR) in order to maximise the detection of reliable changes (Yanofsky and Bickel, 2010). Although proteomics data are typically more variable, none of the reference protein ratios measured in the SOW samples by iTRAQ labelling was significantly variable when these cutoffs were applied, suggesting that the chosen thresholds were robust (Supplementary Figure 2).
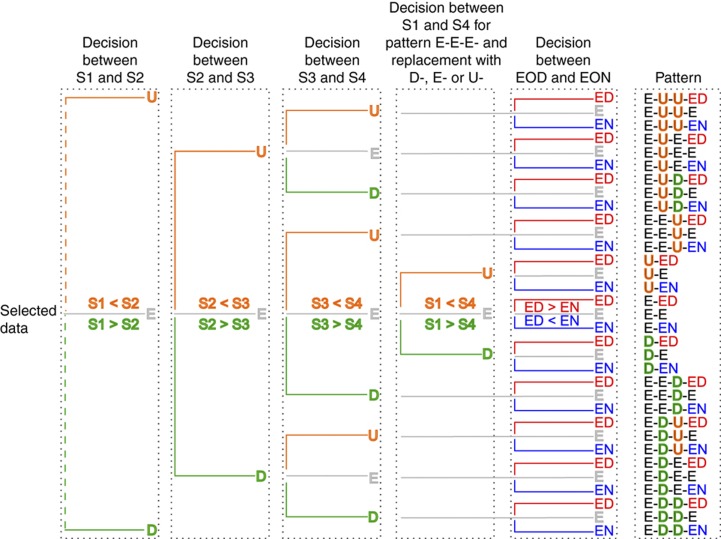
In total, the levels of 17 710 transcripts and 569 proteins changed significantly in SOW samples across growth stages and/or EOD/EON time points, and of 16 370 transcripts and 370 proteins in the equivalent SWD samples. A decision tree was used to cluster similar transcript or protein variation patterns (Figures 5 and 6; Supplementary Figure 3). First, if a transcript or protein level was significantly different (P<0.05) between two successive development stages it was classified as up (denoted ‘U’) or down (denoted ‘D’). For transcripts or proteins whose level was not significantly different between two successive development stages it was denoted ‘E’. For the transcripts and proteins that were initially classified as E-E-E (i.e., no significant change in successive developmental stages), the difference between stages 1 and 4 was also tested so that, in the case of no significant difference, a significant decrease or a significant increase, the E-E-E label was replaced by the corresponding single letter (‘E’, ‘D’ or ‘U’, respectively). Second, the difference between EON and EOD was tested. Here, transcripts and proteins that were higher at EOD were denoted with ‘ED’, those higher at EON with ‘EN’ and those without a significant change between EOD and EOD with ‘E’. Thus, the decision tree theoretically comprises 87 different classifications, and the final label of each classification is the result of each transcript or protein tested in the clustering process. For example, U-D-E-EN represents a pattern in which the abundance ratio increased from stage 1 to stage 2, decreased from stage 2 to stage 3, did not change significantly between stage 3 and 4, and was higher at EON.
Figure 5.
Clustering of transcript and protein profiles with a decision tree. The clustering of the regulated proteins and transcripts into 87 theoretical patterns was done according to the results of the ANOVA. First, the difference between the subsequent stages (stages 1–4=S1–S4) was tested for increasing (orange) or decreasing (green) levels or no significant change (grey). For pattern E-E-E, the difference between stages 1 and 4 was also tested and the E-E-E replaced accordingly with ‘U’, ‘E’ or ‘D’. Then, the difference between EON and EOD was tested for levels higher at EOD (‘ED’, red) or EON (‘EN’, blue) or no significant change (‘E’, grey). Displayed is the subtree for patterns with ‘E’ after the first decision and the full tree is available in Supplementary Figure 3.
Figure 6.
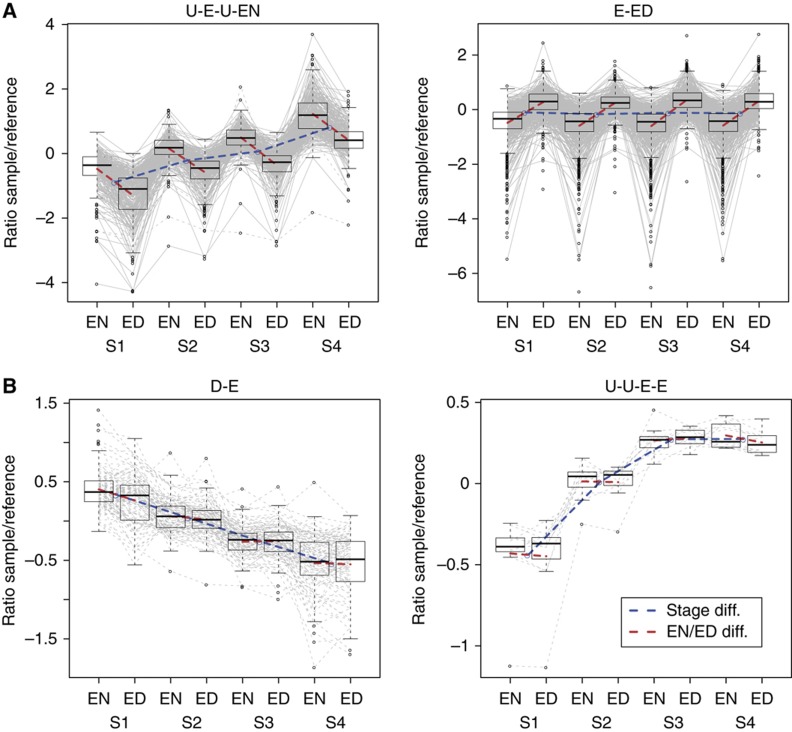
Example transcript and protein patterns. (A) Transcripts in patterns U-E-U-EN and E-ED and (B) proteins in patterns D-E and U-U-E-E. For each transcript or protein in the respective pattern, the mean sample/reference ratios in the eight time points (stages 1–4=S1–S4, EON=EN, EOD=ED) are shown and connected with grey lines. At each time point, a boxplot using Tukey’s standard definition illustrates the distribution of the ratios. The blue line depicts the stage differences by connecting the means between the EOD and EON samples for each stage and the red lines the EOD and EON differences at each stage.
The distribution of transcript and protein patterns (Supplementary Tables 3–7; Supplementary Figures 4–7) was similar between SOW and SWD samples, suggesting that soil water content did not significantly influence the distinction between developmental stages. The transcripts and proteins associated to specific patterns reflected the functional states of the leaf in stage 1 when many cells are still dividing, in stages 2 and 3 when cells are mostly expanding, and in stage 4 when most cells have expanded to their final size and the leaf has reached full photosynthetic capacity (Supplementary Table 8). The marked change in transcript and protein abundance levels between stages 3 and 4 pointed to significant reprogramming once the leaf has reached its full size. Stages 2 and 3, which were qualitatively similar even if differing quantitatively in terms of expansion rate, can be viewed as transition or expansion stages. Their combination led to the identification of protein and transcript expansion stage markers. For the transcripts, the most over-represented GO categories were positive regulation of catalytic activity with mRNAs for four thioredoxins and quinolate synthase, as well as photosynthetic electron transport in photosystem I (Supplementary Table 9). Thioredoxins are known to target photosynthetic proteins in chloroplast thylakoid membranes (Balmer et al, 2006) and cyclic electron flow efficiency around photosystem I has been linked to the assimilatory capacities of leaves before (Breyton et al, 2006). Our results therefore suggest that photosynthesis during leaf expansion is linked to redox control, either for regulation of photosystem complex activity directly or to coordinate photosynthesis with the activity of downstream redox-controlled enzymes.
Diurnal transcript oscillations depend on leaf growth status and are strongly dampened by water deficit
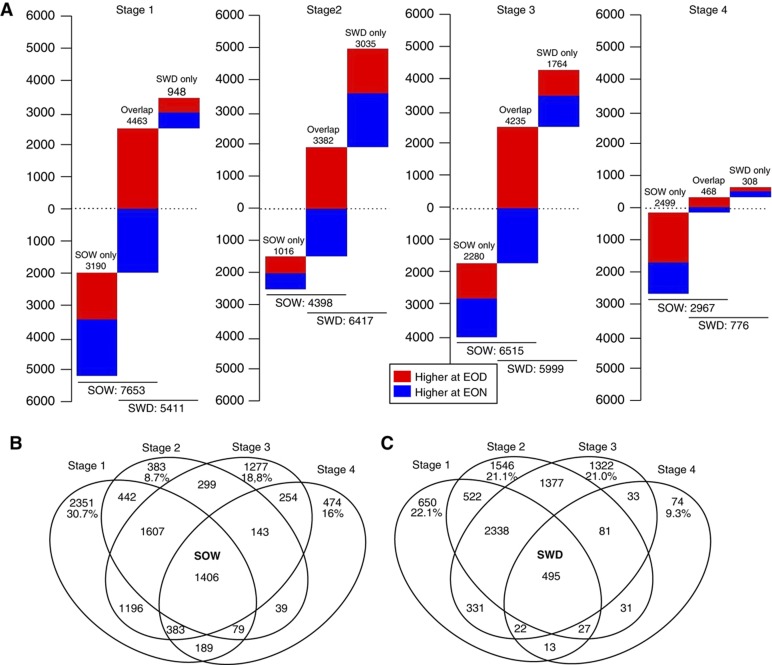
Strong oscillations between EOD and EON were detected for 50.3% of transcripts in SOW and 43.1% in SWD. For example, transcripts of the clock component genes LHY (AT1G01060) and CCA1 (AT2G46830) were higher at EON, while TOC1 (AT5G61380) and GIGANTEA (AT1G22770) were higher at EOD (Alabadí et al, 2001; Locke et al, 2005). Furthermore, the two defence protein genes PHT4;2 (AT2G29650) and ACD6 (AT4G14400) that are induced by light (Lu et al, 2003; Wang et al, 2011) accumulated to higher levels at EOD, but only later during leaf development (Supplementary Figure 8). In addition to the 30–40% of transcripts previously reported to be regulated by the circadian rhythm (Covington et al, 2008), the mRNAs that differed significantly between EOD and EON include those from genes that are diurnally regulated by light and sugar (Usadel et al, 2008) and diurnally regulated transcripts specific for early stages of leaf development that would have been missed at the level of the Arabidopsis full leaf rosette. The number of transcripts that oscillate during the diurnal cycle depended on the growth stages, with markedly fewer transcripts changing in fully expanded leaves (Figure 7A; Supplementary Table 10).
Figure 7.
Stage-dependent diurnal transcript changes. (A) The number of transcripts changing between end-of-day and end-of-night with an adjusted P-value<0.05 in each of the four growth stages. In each stage, the first bar indicates the transcripts that change only in the optimal water experiment, the second bar those that change in both optimal water and water deficit, and in the third bar those that only change in water deficit. The transcripts that are higher at EON are indicated in blue and those at EOD in red. (B) Venn diagram of the stage-dependent diurnal changes in SOW and (C) in SWD. For the transcripts that show diurnal changes only in one stage, the percentage with regard to all transcripts changing in that stage is given.
Fluctuating transcripts with higher levels at EON belong to GO categories significantly over-represented for transcription, while those higher at EOD are enriched for biological processes connected to translation (Supplementary Figure 9). Transcripts in the EOD GO categories showed a typical accumulation in the light that is coordinated with CO2 fixation and the availability of sugars, and therefore decrease during the night in parallel with the gradual metabolism of starch (Usadel et al, 2008). EOD and EON oscillating transcripts are also distinguished by stimulus response pathway GO categories. For example, transcripts higher at EON and associated with GO categories response to abiotic stimulus and gravity include PHYA (AT1G09570), the PHYA interactors PIL5/PIF1 (AT2G20180) and PIL6/PIF5 (AT3G59060), and CRYPTOCHROME1 (AT4G08920), all involved in light perception. Similarly, the PIN3 -(AT1G70940) and AUX1 (AT2G38120) auxin transporters as well as the IAA7 (AT3G23050) auxin-responsive transcription factor are represented in these GO categories (Supplementary Table 11). Although the involvement of light and hormone perception in plant growth regulation is well documented (Nozue and Maloof, 2006), the clustering of the pathway components into different patterns suggests that their relative contribution to leaf growth changes over time, including in the diurnal cycle.
Strikingly, a considerable larger set of genes showed diurnal transcript oscillations during leaf growth in SOW than in SWD conditions except in stage 2 (Figure 7A). This can be explained by the better separation of successive SOW growth stages and the markedly strong oscillations in stage 1 of SOW leaves. In contrast, transitions between growth stages were less sharp in the slower growing SWD leaves (Figure 7B and C). For finding differences in energy allocation processes that might reflect reduced growth in SWD, we compared transcript oscillations for proteins involved in energy allocation processes, e.g., those regulated by sugars (Usadel et al, 2008), in each growth stage between SOW and SWD leaves. All of the reported sugar-induced and sugar-repressed genes were represented in the transcripts that are significantly higher at EOD or EON, respectively, independent of growth conditions, but there were marked differences between the growth stages. Sugar-regulated transcripts were under-represented at all growth stages except in SOW leaf stage 2 and SWD leaf stage 1 (Supplementary Table 12). The responsiveness of gene expression and transcript accumulation to sugar levels therefore depends on growth stage and environmental conditions, suggesting that energy is allocated specifically at different stages of leaf growth. At stage 4, leaf 6 is overgrown and mostly covered by younger leaves. The smaller number of diurnally oscillating transcripts at this stage could therefore be linked to lower light intensity and consequently reduced carbon-fixation and sugar synthesis during the day.
With few exceptions, the measured leaf proteome does not show diurnal oscillations
For the subgroup of transcripts for which we also had quantitative protein levels, we found diurnal transcript oscillations in 74.2% of the genes in SOW, and 73.9% in SWD. In contrast to these large-scale diurnal transcript oscillations and confirming the proteome PCA (Figure 4), only two proteins showed a significant diurnal regulation, CP31A (AT4G24770) and CCL (AT3G26740). The chloroplast 31-kDa RNA-binding protein CP31A is on average 1.7-fold more abundant at EOD in SOW, even though the accumulation of the CP31A transcript is not diurnally regulated. CP31A is required for editing and stability of chloroplast mRNAs and one of several nuclear-encoded RNA-binding proteins involved in the post-transcriptional regulation of chloroplast gene expression (Barkan, 2011). CP31A controls the accumulation of the chloroplast NDHF (ATCG01010) mRNA coding for a subunit of the NADH dehydrogenase complex that regulates the light-dependent reduction of the plastoquinone pool (Tillich et al, 2009). The potential involvement of CP31A in a diurnal process could therefore explain the diurnal oscillation of CP31A. The CCL protein is more abundant at EON but its transcript is more abundant at EOD. CCL is encoded by a highly unstable mRNA in Arabidopsis that is a molecular marker for the circadian rhythm (Gutierrez et al, 2002), although the function of the protein is unknown. CCL mRNA accumulation is controlled by the RNA decay pathway (Lidder et al, 2005). Rapid mRNA turnover at specific times of the day is required for the circadian oscillation of clock-controlled genes (Lidder et al, 2005) and is likely also important for diurnal protein oscillations. The anti-cyclical behaviour of mRNA accumulation and protein abundance also suggests translational regulation of mRNA decay.
The lack of diurnal protein oscillations compared with transcript oscillations has been reported for only a few specific cases, e.g., PHYB (Bognár et al, 1999) and several enzymes of primary metabolism (Gibon et al, 2004). Thus, our discovery that this affects over 1700 quantified proteins throughout leaf growth and development is remarkable. However, we cannot exclude that our finding applies mainly to high-abundance proteins that can be measured in a large-scale proteome analysis and that low-abundance proteins may, at least in part, fluctuate between EOD and EON in correspondence with their transcripts.
Protein and transcript levels are not always positively correlated during leaf growth
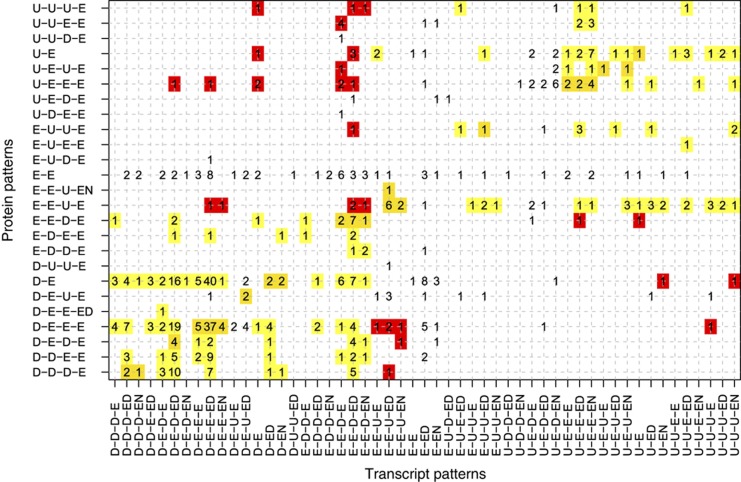
To understand the relationship between the transcriptome and proteome during leaf development, we compared transcript and protein patterns for all 547 genes that had significant changes in both groups. Concomitant down-regulation of protein and transcript levels between growth stages was observed in over 50% of the transcript–protein pairs (Figure 8). However, about 5% (25 pairs) showed opposite trends with decreasing transcript and increasing protein accumulation, including five ATP synthase subunits (average Spearman rank correlation −0.54) and two photosystem subunits (average Spearman rank correlation −0.38) (Supplementary Table 13). All of these 25 proteins are known or predicted components of a membrane system or localised to the plastid (Carbon et al, 2009). In addition, plastid proteins are significantly over-represented in this subset of 25 pairs considering that it includes 19 of 202 protein–transcript pairs (of the 547 total pairs) that are annotated as ‘plastidic’ (P=1.8e−04, Fisher’s exact test) (Baerenfaller et al, 2011). Our results suggest that proteins located in plastids or endomembrane systems can accumulate despite decreasing transcript levels, possibly as a result of post-translational regulatory mechanisms in the trafficking and turnover of membrane-associated proteins, processes that are currently not well understood.
Figure 8.
Correlation of protein and transcript patterns across growth stages. The number of protein–transcript pairs that fall into the respective protein and transcript pattern combinations are given for the protein–transcript pairs in which both the protein and the transcript were changing significantly between the eight time points in the optimal water experiment. Yellow: identical patterns, bright yellow: similar trends, red: opposite trends.
Consistent with the above results, photosynthetic processes are over-represented for proteins that accumulate during leaf growth (U-E-E-E, U-U-E-E, U-U-U-E) while their corresponding transcripts decrease, especially between stages 3 and 4 (E-E-D-E, E-E-D-ED, U-E-D-EN, U-E-D-E). A notable exception is PsbA (ATCG00020), the reaction centre protein of photosystem II that is damaged during photosynthesis. The increase of psbA transcript serves to maintain protein homeostasis despite high turnover of PsbA when PSII activity is high (Mattoo et al, 1989).
Long-term fate of biological processes involved in leaf growth: leaving early developmental processes behind
We performed a meta-analysis of the integrated transcriptome–proteome data and the related GO categories to uncover processes that are important for development and growth of the small young emerging leaf into a fully expanded leaf. Most of the over-represented GO categories linked to development and growth correlate with proteins and transcripts that become significantly down-regulated during leaf growth, especially between stages 1 and 2. Proteins that matched the pattern D-E-E-E belong to GO categories cellular developmental process and cell differentiation and include SMALLER TRICHOME WITH VARIABLE BRANCHES (SVB, AT1G56580), ENOLASE1 (ENO1, AT1G74030) and CDC48 (AT3G09840). SVB and ENO1 both affect trichome development (Marks et al, 2009; Prabhakar et al, 2009). TRANSLATIONALLY CONTROLLED TUMOUR PROTEIN (TCTP, AT3G16640), which also belongs to the same GO categories, best matched the pattern E-E-U-EN (Spearman rank correlation −0.27), with TCTP mRNA levels increasing between stages 3 and 4. Our results confirm that TCTP accumulation is translationally controlled (Brioudes et al, 2010). TCTP protein expression is found in all tissues containing meristematic and expanding cells and has been proposed to regulate plant growth as a mediator of TOR activity (Berkowitz et al, 2008). However, the mRNA levels for proteins in the TOR pathway, TOR (AT1G50030), RAPTOR1A (AT5G01170) and RAPTOR1B (AT3G08850), did not change significantly during leaf growth. The GO category organ development was also over-represented for proteins down-regulated between stages 1 and 2 (D-E-E-E). Together, our results suggest that proteins important in leaf development act early and become rapidly down-regulated before cell expansion.
Translation and protein metabolism are prevalent processes during early leaf growth
Other proteins in the GO category organ development include HSP90.2 (AT5G56030), HSP90.7 (AT4G24190), RACK1B (AT1G48630) and RACK1C (AT3G18130), which are stage 1 markers involved in protein metabolism. Protein levels for the third RACK1 homologue RACK1A (AT1G18080) also decreased across growth stages (D-D-E-E) and all three RACK1 proteins showed a good correlation between transcript and protein levels (average Spearman rank correlation 0.49). RACK1 has been implicated in plant development (Chen et al, 2006) and abscisic acid response, and was shown to interact with eIF6, a regulator of ribosome assembly (Guo et al, 2011). The HSP90 chaperone complex participates in protein folding and its inhibition also affects leaf development (Sangster and Queitsch, 2005). HSP90 consists of seven isoforms, which are located in the cytoplasm (HSP90.1–HSP90.4), plastid (HSP90.5), mitochondria (HSP90.6) and the endoplasmatic reticulum (HSP90.7) (Krishna and Gloor, 2001). In addition to HSP90.2 and HSP90.7, the protein level of HSP90.5 (AT2G04030) declined during development (D-D-D-E), while HSP90.6 (AT3G07770) protein levels did not change significantly. Together with RACK1 and HSP90, other proteins involved in translation and protein folding have been identified in the set of down-regulated stage 1 marker proteins, including two TCP-1/cpn60chaperonin family proteins (AT1G24510, AT5G26360), NACA2 (AT3G49490), NACA3 (AT5G13850) and ribosomal proteins. Thus, the prevalence of translation and protein metabolism in emerging leaves is also well reflected in the stage 1 marker proteins (Supplementary Table 5).
Dynamic changes in ribosomal transcripts and proteins during leaf growth
Ribosomal proteins accumulate to high levels in proliferating cells (McIntosh and Bonham-Smith, 2006). Protein- and transcript-level data are available for 110 ribosomal proteins in our complete data set. Among these, 107 transcripts were significantly down-regulated in at least one of the four leaf growth stages but never up-regulated, while 49 proteins also declined (average Spearman rank correlation=0.52) and 61 did not show a significant change (average Spearman rank correlation=0.29). Genes coding for subunits of constitutive protein complexes such as the ribosome show significant transcriptional co-regulation in yeast, but it was postulated that co-regulation of subunits of a protein complex should be detectable primarily at the level of protein abundance (Jansen et al, 2002). The absence of significant abundance changes for nearly 60% of the measured ribosomal proteins therefore suggests that regulatory mechanisms ensure protein homeostasis despite decreased transcript levels, possibly through stabilisation of individual ribosomal proteins once the ribosome has been assembled.
The patterns of all 349 clustered transcripts coding for ribosomal proteins in Arabidopsis followed the trend discussed above, i.e., 317 transcripts were significantly down-regulated in at least one of the leaf growth stages and were never up-regulated. Only 14 ribosomal protein mRNAs increased during leaf growth and did not decline (Supplementary Table 14). Interestingly, five of these 14 ribosomal protein mRNAs encode L18a family members (four members of the ribosomal protein LA18ae family and the 60S ribosomal protein L18A-1). The specific functions of the L18a proteins are not known, but our data show that members of the L18a and L18ae/LX subfamily are differently regulated and that they may be assembled into ribosomes only late during leaf growth, possibly replacing other subunits that function earlier in development. Consistent with the over-representation of transcripts higher at EOD in the GO category translation, 270 of the 349 ribosomal protein transcripts have higher levels at EOD. Interestingly, of the 30 transcripts that have higher levels at EON, eight also belong to the above group of 14 transcripts that were up-regulated during leaf growth (over-representation with P-value 1.3e−05, Fisher’s exact test), including the L18a transcripts. This suggests again that specific members of the L18a ribosomal protein genes are regulated differently and therefore may have specific functions during leaf growth.
Cis-regulatory elements that are over-represented in the promoters of the 353 nuclear-encoded ribosomal genes were identified using ATCOEcis (Vandepoele et al, 2009). They include the telo-boxes (AAACCCTA, P-value 2.4e−41; AAACCCTAA, P-value 7.2e−21) and the site II element (TGGGCY, P-value 2.1e−15) related to ribosomal protein gene expression (Trémousaygue et al, 2003) (Supplementary Table 15), as well as the PHYA-responsive SORLIPs 2 motif (GGGCC, P-value 1e−16) that is a light-responsive cis-regulatory sequence (Hudson and Quail, 2003). An over-representation of light-dependent promoter elements was not observed for other groups of transcripts that are higher at EON or EOD. Our integrated large-scale data therefore establishes that ribosomal proteins are closely co-regulated during leaf development and growth through specific promoter cis-regulatory elements.
Senescence- and autophagy-associated transcript patterns already emerge during leaf expansion
Specific gene expression patterns that are diagnostic of the mature and senescing leaf were already discernible during leaf expansion. For example, the transcript level of aleurain-like protease (AT5G60360) that is up-regulated during senescence (Gepstein et al, 2003) continuously increased during leaf growth (U-U-U-EN), while up-regulation of the protein was only detected between stage 3 and 4 (E-E-U-E). Similarly, transcripts that accumulated significantly between stage 3 and 4 (U-U-U-EN, E-E-U-EN and E-E-U-E) were enriched for the GO category aging, and include the senescence-associated genes SAG12 (AT5G45890), SAG13 (AT2G29350), SAG20 (AT3G10985), SAG101 (AT5G14930) and SRG1 (AT1G17020). Autophagy is a key metabolic process during leaf senescence. The transcript patterns for the 28 autophagy-related genes in Arabidopsis revealed that 25 were higher at EON, and 22 significantly increased during at least one-stage transition (Supplementary Table 16). Autophagy appears to be initiated before completion of leaf expansion (Breeze et al, 2011), and our results that the autophagy-related genes ATG2, ATG5, ATG8D, ATG8F, ATG8H/I, ATG12A and ATG18G were already significantly up-regulated during the transition from stage 1 to 2 support this view. ATG7, ATG8A, ATG8B and ATG8H were reported to be up-regulated just before the onset of senescence and ATG7 was proposed to be the key control point for autophagy activation (Breeze et al, 2011). Our results show that expression of ATG8A, ATG8B and ATG8D was already significantly up-regulated during the transition from stage 3 to 4 (E-E-U-EN), while ATG7 transcript levels did not change significantly during leaf growth (pattern E-EN). Although the stage 4 leaf was not visibly senescent, interestingly the senescence- and autophagy-associated transcriptional programs were triggered during leaf growth, while expression of the proposed key regulator ATG7 remained unchanged.
Cell wall modification signatures during leaf growth reflect cell expansion and pathogen resistance
In growing tissues, plant cells expand massively to reach their final shape and size while resisting turgor pressure. Cell expansion is tightly controlled through remodelling of the cell wall cellulose–matrix network (Szymanski and Cosgrove, 2009), and cellulose synthesis is largely confined to expanding cells (Somerville et al, 2004). Furthermore, cell wall loosening is necessary for anisotropic growth because it determines which cell walls must yield under stress (Szymanski and Cosgrove, 2009). Cell expansion and anisotropic growth are reflected in our data set by genes for proteins associated with cell wall biogenesis, organisation and loosening that were over-represented in patterns of decreasing transcript levels (E-D-E-EN, E-D-E-E, E-E-D-EN, E-E-D-ED). Genes in this class included cellulose synthase (CESA), expansin and expansin-like proteins (Supplementary Table 17). Pectin methyl esterases are thought to be secreted late during leaf growth (Szymanski and Cosgrove, 2009), stiffen the pectin gel and reduce cell growth. We found, however, that the transcripts coding for the pectin methylesterases ATPME1 (AT1G53840, E-E-D-ED), ATPME3 (AT3G14310, E-E-D-ED), PME61 (AT3G59010, D-D-D-ED) and ATPME44 (AT4G33220, U-U-D-ED) were down-regulated between stages 3 and 4. Thin-walled cells were found to grow faster than thick-walled cells (Refrégier et al, 2004). Correspondingly, the GO category cell wall thickening was over-represented by transcripts increasing during leaf growth (U-U-E-ED, U-E-E-E). Transcripts that contribute to the over-representation in these patterns include those for cytochrome P450 polypeptides and UDP-glycosyltransferase 74B1 (AT1G24100), which is involved in the synthesis of glucosinolates from tryptophan. ASA1 (AT5G05730) regulates the defence-dependent synthesis of indole glucosinolates (Clay et al, 2009), which is consistent with the transcriptional up-regulation of ASA1 during leaf growth. The two up-regulated genes PEN2 (AT2G44490) and PEN3 (AT1G59870) have also been implicated in resistance to pathogens (Clay et al, 2009) and are required for callose deposition and glucosinolate activation. Together, the transcript patterns at the four leaf growth stages significantly expand information on the expression of genes for proteins involved in cell wall biogenesis, cell wall loosening and cellulose deposition enabling cell expansion. The cell walls thicken only later during leaf growth, thereby inhibiting cell expansion and building up a barrier against pathogen attack.
Chloroplast gene expression during leaf growth involves a switch between nuclear- and plastid-encoded RNA polymerases
In contrast to the over-representation of photosynthetic processes among transcripts decreasing during leaf growth, transcript levels of several plastid genes coding for subunits of the photosynthetic complexes were increasing (Supplementary Table 18). Chloroplast genes are transcribed by two single-subunit nuclear-encoded RNA polymerases (NEPs) and a multi-subunit plastid-encoded RNA polymerase (PEP). NEP appears to be mainly active during chloroplast development and in transcribing genes with housekeeping functions, while PEP is the principal RNA polymerase in the mature chloroplast and responsible for transcribing photosynthesis-related genes (Liere et al, 2011). Consistent with the switch from NEP to PEP activity, levels of NEP-related transcripts decrease during leaf growth (Supplementary Table 18). Thus, despite the complexity of post-transcriptional RNA processing in chloroplasts (Barkan, 2011), most of the regulation of chloroplast transcript abundance during leaf growth can be explained by changes in transcriptional activity. PEP activity is regulated by nuclear-encoded sigma-type transcription initiation factors (Lerbs-Mache, 2011). Transcript levels for SIG2 and SIG6, which are the two essential sigma factors for chloroplast functions, decreased significantly between stage 3 and 4 and were higher at EOD (pattern E-E-D-ED). In contrast, the mRNA level for SIG5, which accumulates in the light and predominantly binds to the psbA and psbD promoters (Onda et al, 2008), was up-regulated significantly between stages 1 and 2 and was higher at EON (U-E-E-EN). The regulation of individual sigma factors during leaf growth therefore suggests that chloroplast genes continue to be differentially transcribed even after the switch to PEP transcription.
Water deficit adaptation reduces the expression of genes supporting fast growth
The direct comparison of SOW and SWD leaf 6 data revealed 1222 (5%) differently regulated transcripts (P<0.05 in a paired t-test, average fold-change >1.5) (Supplementary Table 19). The third principal component of the combined transcript data PCA analysis separated the growth stages in the individual datasets, but did not separate between SOW and SWD samples, indicating that the growth stages are comparable between the two experiments (Supplementary Figure 10) and that their definition based on growth variables was pertinent (Figure 1). Among the differently regulated transcripts, 368 transcripts that accumulated to higher levels in water deficit are over-represented in the GO categories carbon-fixation and response to metal ion. This result is consistent with previous reports that leaves under water deficit have an increased turnover and availability of C metabolites (Hummel et al, 2010; Skirycz et al, 2010; Tardieu et al, 2011). Consequently, reduced growth in water limiting conditions is not a consequence of a decrease in fixed carbon, but rather an adaptation response. The 854 transcripts that accumulated at higher levels in SOW leaves were most enriched for the GO categories ribosome biogenesis, translation and defence response to fungus. The first two GO categories are consistent with the higher biosynthesis activity required to support the growth rate of SOW leaves (Figure 1).
Water deficit adaptation differs significantly from a drought stress response
Transcripts that account for the over-representation of the GO category defence response to fungus include OCP3 (AT5G11270), the peroxidase superfamily protein PEROXIDASE CB (AT3G49120), PEROXIDASE SUPERFAMILY PROTEIN (AT5G64120), PR4 (AT3G04720), MLO12 (AT2G39200) and ETHYLENE RESPONSE FACTOR 104 (AT5G61600) (Supplementary Table 20). Transcription of peroxidases and other genes indicative for oxidative burst is induced upon biotic stimulus as rapid generation of superoxide and accumulation of H2O2 characterise the hypersensitive response (Lamb and Dixon, 1997). Because reactive oxygen species also increase upon drought stress as by-products of drought stress metabolism and as signalling molecules (Achard et al, 2006; Cruz de Carvalho, 2008; Miller et al, 2010), these genes would have rather been expected to be more highly expressed in SWD leaves if they displayed a drought stress response. The same reasoning also applies to the expression of genes encoding biotic defence proteins and ethylene response factors, which increase in response to abiotic stress (Navarro et al, 2008; Skirycz et al, 2010). Therefore, we investigated whether previously reported water-deficit stress marker genes were induced in SWD leaves. The results in Supplementary Table 21 show that transcript levels of genes involved in the biological processes affected by osmotic stress (Skirycz et al, 2010) were either not significantly different between SOW and SWD leaves or in some cases even lower in SWD leaves. Transcripts that accumulated to significantly higher levels in SOW leaves include the biotic stress markers MLO12 (AT2G39200) and PR5 (AT1G75040), as well as CYP71A13 (AT2G30770) and CYP57220 (AT5G57220) that are involved in indole glucosinolate biosynthesis. DELLA proteins restrict growth in adverse growth conditions and integrate the ABA, GA and ethylene signalling pathways in response to both salt and drought stress (Achard et al, 2006; Skirycz et al, 2010). ABA-independent signalling pathways that activate the expression of DREB/CBF transcription factors are also induced upon stress treatment (Shinozaki and Yamaguchi-Shinozaki, 1996; Harb et al, 2010). Our results show, however, that none of the genes involved in hormone synthesis and signalling that have been implicated in regulatory responses to abiotic stress conditions were expressed at significantly higher levels in SWD compared with SOW leaves (Supplementary Table 21).
Interestingly, transposable elements (P-value=7.2e−11) and pseudogenes (P-value=2.4e−9) are strongly over-represented in the group of transcripts that differed between SOW and SWD leaves. While pseudogenes and transposable elements are transcriptionally activated during stress (Zeller et al, 2009), these transcript categories are more over-represented in SOW (pseudogenes, SOW P-value=2.1e−7 and SWD P-value=1.3e−3; transposable elements, SOW P-value=2.2e−16 and SOW P-value=9.9e−4).
Together, the transcriptional response in SWD plants growing in soil in which the water content was reduced early and kept at a constant reduced level during development and leaf growth is significantly different from transcriptional responses in plants exposed to a sudden water stress. Our results show that plants adapt to a low soil water potential by adjusting their growth and gene expression, possibly to avoid an acute water deficit.
Adaptation to water deficit is associated with up-regulation of cold-induced proteins
Analysis of the proteomes of the leaf 6 growth stages in SOW and SWD leaves revealed 34 differentially regulated proteins (P<0.05, average fold-change >1.5) (Supplementary Tables 22 and 23). Of these, 18 accumulated to higher levels in SOW leaves, mainly comprising proteins involved in translation, corresponding to the trend observed in the transcriptome data. The 16 proteins significantly more abundant in SWD leaves included COR15A (AT2G42540), COR15B (AT2G42530) and COR6.6/KIN2 (AT5G15970), all known to be expressed in response to cold. Their induction can be explained because water availability to plant cells may be limited by distinct but functionally related abiotic stresses (Verslues et al, 2006) including low soil water potential during drought and in high saline soil conditions, but also cellular dehydration at low temperature as a consequence of ice crystal formation. Avoidance and tolerance responses to dehydration are in part mediated by the cold-induced transcription factor DREB1A/CBF3 (AT4G25480) and the drought/high salinity-induced transcription factor DREB2A (AT5G05410) whose target genes partially overlap (Sakuma et al, 2006) (Supplementary Table 21). Interestingly, expression of COR15A, COR15B and COR6.6/KIN2 is specifically activated by DREB1A/CBF3 in response to cold (Sakuma et al, 2006). However, the transcript levels for DREB1A/CBF3, COR15A, COR15B and COR6.6/KIN2 did not differ between SOW and SWD leaves, and were higher for DREB2A in SOW leaves (P-value<0.05) without meeting the fold-change cutoff. Consistent with the other stress markers discussed above, SWD plants did not trigger a stress-related transcriptional activation of the DREB1A/CBF3, COR15A, COR15B and COR6.6/KIN2 genes but adapted to the reduced soil water content. Interestingly, this adaptation involved the increased accumulation of certain stress response-related proteins, but their regulation under a continuous abiotic stress could clearly be distinguished from the induced transcription of their genes that typically occurs in response to short-term water-deficit conditions.
Discussion
The comprehensive phenotypic and quantitative molecular profiling data presented here significantly expand our knowledge about the systems function of a developing leaf through successive stages of development in two different water regimes. We used Arabidopsis leaf number 6 as a model because its growth phenotypes were previously analysed in other genotypes both in optimal watering condition (Cookson et al, 2005; Massonnet et al, 2010, 2011) and in response to reduced soil water content (Cookson et al, 2006; Tisné et al, 2010), which were the conditions used in our experiments. Reduced soil water content, which plants may experience during prolonged drought, markedly affects leaf growth. In our condition this resulted mainly in a reduced final leaf area and, to a lesser extent, reduced final leaf thickness. The reduced leaf growth was mostly due to decreased expansion rates in the two dimensions and was partly compensated by an extended growth period (Aguirrezabal et al, 2006; Skirycz and Inzé, 2010). Related to reduced leaf growth we also measured a reduction in DNA ploidy in SWD leaves, but only after their rapid phase of expansion. Reduction in the number of DNA endoreplication cycles per cell was also reported for mature leaf 6 in Col-0 and different mutants (Cookson et al, 2006), but not in leaf 3 (Skirycz et al, 2010), suggesting that DNA ploidy is tightly controlled in different environmental conditions later during plant development. As detailed above, our comprehensive molecular profiling results, particularly the quantitative proteome analysis, similarly support and extend previously reported results for leaf growth, thus underpinning the high quality of our data.
Our experimental approach also provides new biological insights into the molecular mechanisms governing leaf growth. We found that transcript and protein variation patterns reflect the functional state of the leaf at each growth stage. The majority of protein–transcript pairs across the four leaf growth stages correlate well with some interesting exceptions, which indicate that subcellular protein localisation and complex formation also determine protein-level regulation. Unexpectedly, we discovered that a large number of transcripts show strong diurnal fluctuations that are not matched by corresponding protein-level fluctuations at the experimental time scale (8 h between EON and EOD). Because MS-based proteomics is inherently biased towards more abundant proteins, we considered that this could have introduced a technical bias in our data against the detection of diurnal protein-level changes. In the subset of transcripts for which we also had quantitative protein data, however, we found significant abundance changes for 74% of the transcripts, while in the detected proteome we found diurnal changes only for two proteins. Also, the reported underestimation of ratios in iTRAQ quantitation (Karp et al, 2010) cannot explain our results, because the underestimation affects high ratios and we were able to detect changes of up to nearly 12-fold in our iTRAQ data. In addition, when combining significance testing with a fold-change cutoff we could set the fold-change cutoff to only 1.5. This allowed us to detect small but reliable changes, even if the rigorous statistical tests also resulted in some false negatives. Moreover, we detected significant abundance changes in an unusually large fraction of the identified proteins in SOW (27.4%) and SWD (24.5%) leaves. Thus, the nearly absent protein-level fluctuation between EOD and EON in the detected proteome most probably points to currently unknown regulatory mechanisms controlling protein levels, such as post-translational processes that modulate protein homeostasis by feed-back loops between protein and transcript levels and targeted protein degradation. Our results in Arabidopsis are reminiscent of the observed post-transcriptional noise in yeast that buffers protein levels against mRNA fluctuations and was estimated to affect at least 25% of the proteome (Lee et al, 2011). The reduced diurnal protein-level variation could also be explained, at least in part, by the relative concentrations of proteins and transcripts and different time constants for transcript and protein synthesis and degradation. For example, for enzymes of central metabolism the relative amount of transcript to protein is so low that in a leaf it may take several days for a change in transcript level to cause a major change in protein level (Piques et al, 2009). A recent study in mammalian cells showed that proteins are on average about 900 times more abundant than their corresponding transcripts and the energy consumed for the production of proteins is nine-fold larger than that for transcripts (Schwanhäusser et al, 2011). Thus, it requires considerably more catabolic or anabolic activity to produce a significant change in protein versus transcript levels. Consequently, the high cost of protein synthesis may justify on its own that, by default, cells prefer a relatively slow protein turnover in leaves as well. But then, the interesting question remains why 74% of the transcripts in the transcriptome subset for which we have proteome data show diurnal fluctuations throughout growth if this has little impact on protein abundance. Transcript fluctuations might prime cells for a faster response to stress or changing environmental conditions, which would be more difficult to achieve at constant mRNA concentration. The general dampening of mRNA-level oscillations in SWD is consistent with this view and suggests that plants can also adjust overall mRNA metabolism to a continuous suboptimal growth condition. Together, the dampened diurnal fluctuation of moderately to highly abundant proteins in leaves detected in our mass spectrometry analysis might result in energy savings in stable conditions due to low protein turnover rates, while significant transcript fluctuation might enable rapid reprogramming to respond to environmental changes. This does not exclude that although fluctuating, the observed levels of certain mRNAs at specific times during the diurnal cycle may be required to regulate the energy status of the cell at that particular time, the respective growth stage, or in the prevailing environmental condition. This could also explain the differences in growth stage- and experiment-dependent diurnal transcript oscillations.
Finally, plants grown in soil with continuous reduced water potential were shown to exhibit a systems-level adaptation process, which is substantially different from the well-established response to drought stress. Our results allow us to distinguish between direct large-scale effects resulting from experimental treatments and secondary effects imposed by the developmental program underlying leaf growth. Our comparative analysis of transcriptome, proteome and phenotypic changes occurring in the leaf in stable and controlled experimental conditions expanded our understanding of systems-level processes in leaf growth and therefore provides the necessary basis for the correct interpretation of results from studies of mutations or stress treatments.
Materials and methods
Plant material and growth conditions
Six successive experiments (Exp. 1 to Exp. 6) were carried out using seeds of the A. thaliana accession Col-4 (N933) obtained from a single batch provided by the Nottingham Arabidopsis Stock Centre. For each experiment, 504 pots were filled with a mixture (1:1, v/v) of a loamy soil and organic compost at a humidity of 0.30 g water per gram dry soil. Ten millilitres of a modified one-tenth-strength Hoagland solution was added to the pot surface just before sowing. Six seeds were sown dispersed over the pot surface. The pots were transferred to a growth chamber equipped with the PHENOPSIS automaton (Granier et al, 2006). After 2 days in the dark, day length in the growth chamber was fixed at 8 h using a mix of cool-white fluorescent tubes, sodium and hydrargyrum quartz iodide (HQI) lamps. The other growth conditions were as follows: air temperature at 21.2°C during the day and 20.5°C during the night; air humidity of 70%; and incident light measured at the plant level ∼220 μmol/m2/s. During the germination phase (until stage 1.02; Boyes et al, 2001), water was sprayed at regular intervals on the pots to maintain sufficient humidity at the soil surface. Beginning at plant germination, each pot was weighed twice a day to calculate the soil water content. For the optimal water condition experiment, the soil water content was adjusted to 0.40 g water per gram dry soil and for the water-deficit experiment plants were in mild water-deficit conditions with soil water content adjusted to 0.24 g water per gram dry soil. The adjustment was done automatically with the PHENOPSIS automaton by addition of an appropriate volume of nutrient solution. Profiling data were obtained from the same biological samples except for stage 1 leaves of the first biological replicate for which protein and transcript data were obtained from different, but identically grown batches of leaves.
Leaf growth measurements
Leaf area
From stages 1.2 to 6.0, five rosettes per genotype were dissected every two to three days. The leaf-6 area (mm2) was measured with image-analysis software (Bioscan-Optimas version 4.10) after imaging with a binocular magnifying ( × 160) glass for leaves smaller than 2 mm2 or with a scanner for larger ones.
Leaf thickness, cell density and volumes
From stage 1.02–6.00, five plants were collected every 2–3 days and whole seedlings (when 6th leaf <4 mm) or leaves were fixed, conserved and subsequently cleared and stained using propidium iodide as described by Wuyts et al (2010). Image stacks covering the complete leaf thickness were produced for the central part of the leaf along the longitudinal axis, and approximately midway between the leaf midvein and margin using multiphoton laser scanning microscopy (Wuyts et al, 2010). The quantitative analysis of leaf thickness, cell density and dimensions in image stacks involved specifically developed ImageJ macros ( http://rsb.info.nih.gov/ij/) and R scripts (R Development Core Team, 2010).
Area and number of epidermal cells
A negative imprint of the adaxial epidermis of the same sixth leaf for which also the surface was measured was obtained after evaporation of a varnish spread on its surface. These epidermal imprints were analysed using a microscope (Leitz DM RB; Leica) supported by the image-analysis software Optimas. Mean epidermal cell density (cells/mm2) was estimated by counting the number of epidermal cells in two zones (at the tip and base) of each leaf. Total epidermal cell number in the leaf was estimated from epidermal cell density and leaf area. Mean epidermal cell area (μm2) was measured from 25 epidermal cells in two zones (at the tip and base) of each leaf.
Estimation of dynamic variables
Leaf area, leaf thickness, epidermal cell number and epidermal cell area were plotted as a function of time (days after leaf initiation). Leaf initiation was estimated when the leaf area was 0.001 μm2. Sigmoid curves Equation (1) were fitted to the data to estimate the rate of processes at each stage.
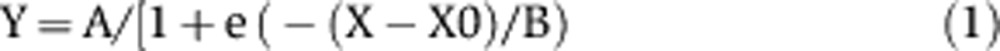 |
This gave an increase in leaf area with A=88.1, B=2.6 and X0=15.8 in SOW and A=57.9, B=4.1 and X0=21.6 in SWD, leaf thickness with A=206, B=6.4 and X0=11.7 in SOW and A=161.9 B=5.0 and X0=12.7 in SWD, cell number with A=26 738.0, B=2.8 and X0=8.4 in SOW and A=20 338.8, B=2.8 and X0=9.8 in SWD, and cell area with A=3478.4, B=3.6, X0=15.7 in SOW and A=2966.9, B=3.6 and X0=19.3 in SWD (Figure 1). Based on leaf-6 area, thickness, epidermal cell number and epidermal cell area changes with time, the four stages were identified to harvest samples for molecular profiling.
Harvests for molecular profiling
Stages of rosette development for all the plants grown in the PHENOPSIS platform were noted every 2–3 days during the six experiments. Based on the first experiment in each environmental condition (Exp.1 and Exp.4), stages of rosette development when the leaf 6 reached the four stages were identified and were used in the following experiments to reproduce a similar sample between experiments. The day before leaf harvest, plants to be harvested were marked with a small plastic tag, pointing toward the tip of the 6th leaf. At each stage, leaves 6 were harvested during the last hour before the light is on (under green light) and before the light is off. To collect enough leaf material for profiling with the different technologies, each sample was prepared by bulking material from numerous plants. The frozen plant material was sent to the MPI in Golm, where it was ground and aliquoted using a cryogenic grinder instrument (German Patent No. 08146.0025U1).
DNA ploidy level
At each stage and under each water condition, the sixth leaf was collected from five plants and was frozen individually and immediately in liquid nitrogen. Flow cytometry analysis was done as described by Cookson et al (2006). For each sample, 3000 nuclei were counted and the percentage of cells at 2C, 4C, 8C, 16C, 32C and 64C was calculated. The endoreduplication factor (i.e., the mean number of endocycles per 100 cells) was calculated from these percentage values as follows: EF=0 × %2C+1 × %4C+2 × %8C+3 × %16C+4 × %32C+5 × %64C.
Tiling array transcript data
Gene expression in leaves of the four developmental stages and at the two diurnal time points in the short-day optimal water (SOW) and short-day mild water-deficit (SWD) experiments, as well as in a reference mixed rosette sample was profiled using AGRONOMICS1 microarrays (Rehrauer et al, 2010) matched against the TAIR10 CDF file. After exclusion of the probe sets for plastid transcripts, 25 150 probe sets representing 24 985 genes gave above-background signals with P-value<0.05 in at least one of the samples in SOW and 27 882 transcripts representing 27 707 genes in SWD. The log2-transformed sample/reference ratios were used in all the analyses. A more detailed explanation is provided in the Supplementary Experimental Procedures. Microarray raw and processed data are available via ArrayExpress (E-MTAB-1056) and the AGRON-OMICS data repositories.
Quantitative RT–PCR transcript data for plastid transcripts
Since most plastid mRNAs are not polyadenylated and the labelling for the AGRONOMICS1 microarray was based on oligodT primers, the transcripts for 80 plastid-encoded genes were profiled for the 24 SOW samples and the reference sample with quantitative RT–PCR (RT–qPCR). Again, all the analyses were based on the log2-transformed sample/reference ratios. A comparison of the standard deviations of the replicate means for the microarray and RT–qPCR transcript profiles for the 69 plastid transcripts for which we have both data types confirmed that the RT–qPCR data are more robust, as their average standard deviation was 0.27, versus 0.58 for the microarray plastid transcript data (Supplementary Figure 11). For this reason, only the RT–qPCR plastid transcript data were used in all further analyses.
Quantitative iTRAQ proteomics data
Proteins in leaves of the four developmental stages and at the two diurnal time points in the SOW and SWD experiments and of the reference sample were quantified using the 8-plex iTRAQ isobaric tagging reagent (Ross et al, 2004; Pierce et al, 2008) according to the labelling scheme in Supplementary Tables 24 and 25. The labelled peptides were fractionated with strong cation-exchange (SCX) and purified before mass spectrometry measurements on an Orbitrap mass spectrometer. The resulting spectra were matched to peptides with the database-dependent search algorithm Mascot (Mascot Science, London, UK) searching the TAIR10 protein database (Lamesch et al, 2012) and the peptide spectrum assignments were filtered for peptide unambiguity in the pep2pro database (Baerenfaller et al, 2011; Hirsch-Hoffmann et al, 2012). Accepting only unambiguous peptides with ion score >24 and an expect value <0.05 resulted in 203 158 assigned spectra at a spectrum FDR of 0.09% in SOW and 145 564 assigned spectra at a FDR of 0.1% in SWD. In SOW, quantitative information for all reporter ions was available in 144 538 of these spectra leading to the quantification of 2081 proteins based on 8710 distinct peptides (Supplementary Table 26). In SWD, 1509 proteins were quantified based on 5292 peptides and 74 550 spectra (Supplementary Table 27). The histograms of the log2-transformed sample/reference protein ratios in the different samples of biological replicate 1 in SOW demonstrate that the ratios were about normally distributed and display similar variances (Supplementary Figure 12), which allowed for the statistical analyses and the comparisons detailed below. A PCA (Figure 4) and correlation analysis (Supplementary Table 28) were performed as quality control and to assess the variance in the data. A more detailed explanation is provided in the Supplementary Experimental Procedures. The proteomics data are available from the PRIDE database (Vizcaíno et al, 2010) (accessions 21330–21353).
Statistical analyses and clustering of the protein and transcript changes
The statistical analytical methods were identical for the protein and transcript data. For each individual data set, the log2-transformed sample/reference ratios were subjected to an analysis of variance (ANOVA) treating stage (S) and day-time (ND) as main effects. The resulting P-values for the global F-test, the stage-dependent level changes and the day-time-dependent level changes were corrected for multiple testing with the Benjamini–Hochberg method (Benjamini and Hochberg, 1995) controlling the FDR to give pGlobal (P-value for an overall global change), pS (P-value for a change between stages) and pND (P-value for the diurnal change). The effect size of the individual stages and the significance of the level changes were computed with the Tukey Honest Significant Differences (TukeyHSD) post-hoc test followed by correction with the Benjamini–Hochberg method (Supplementary Table 29).
Only transcripts and proteins with a pGlobal<0.05 and a maximum fold-change>log2(1.5) were considered for further analysis. In addition, proteins had to have at least one value in each of the eight time points, because a value for each pairwise comparison was an absolute prerequisite for the clustering. This excluded 30 proteins in SOW and 16 in SWD from clustering (Supplementary Table 30). Clustering of the significantly changing proteins resulted in 25 populated clusters in SOW and 19 in SWD and for the transcripts, 77 in SOW and 68 in SWD (Supplementary Tables 3–7; Supplementary Figures 4–7). For a small subset of genes, the protein and transcript variation patterns were verified using qRT–PCR and western blotting (Supplementary Figures 13 and 14; Supplementary Experimental Methods).
We defined protein and transcript stage markers by selecting those that were significantly different in one stage, but not between the other stages (e.g., a stage 1 marker has: P-value stage 1–stage 2 <0.05 and P-value stage 1–stage 3 <0.05 and P-value stage 1–stage 4 <0.05 and P-value stage 2–stage 3 ≥0.05 and P-value stage 2–stage 4 ≥0.05 and P-value stage 3–stage 4 ≥0.05) (Supplementary Tables 4, 5 and 8). For the integration of the proteomics and transcriptomics data, the protein and transcript patterns that occurred in both data sets were combined. This resulted in 220 different combined groups in SOW and 152 in SWD.
For assessing the stage-dependent diurnal changes, a two-sided Welch test was performed and the resulting P-values were corrected for multiple testing with Benjamini–Hochberg. In the correlation analysis of the protein–transcript pairs, the Spearman’s rank correlation coefficients were calculated and P-values were generated with a t-statistic approach corrected with Benjamini–Hochberg.
The comparison of the protein and transcript levels between the SOW and SWD experiment was performed with a paired t-test comparing the values for the eight time points between the two experiments corrected with Benjamini–Hochberg.
GO functional classification
Assignment of protein and transcript functional categories was based on the TAIR GO categories from aspect biological process (ATH_GO_GOSLIM_20110301.txt) excluding annotations inferred from electronic annotation (GO evidence code IEA). The assignment was performed in R (R Development Core Team, 2010) using the elim algorithm from the topGO package (Alexa et al, 2006). Over-representation of categories was assessed using Fisher’s exact test. For each set of AGIs that was subjected to the assessment of over-representation the GO categories with P-value<0.01 were kept and assigned to a list of selected higher-level GO terms according to the GO graph structure.
Supplementary Material
Supplementary figures S1-14, Supplementary tables S1-30
Acknowledgments
We thank the Functional Genomics Center Zurich for providing infrastructure and technical support, Sarah Gerster from the Seminar of Statistics at ETH Zurich for additional statistical support and Johannes Fütterer from the Department of Biology at ETH Zurich for critical reading of the manuscript. This work was supported by the AGRON-OMICS integrated project funded in the sixth European Framework Programme (LSHG-CT-2006-037704).
Author contributions: KB, CM, SW, RS, SB, LH, IS, MS, PH, CG and WG contributed to conception and design of the experiment; KB, CM, SK, KH, AR, DRussenberger, DRutishauser, JS, NW and LH to the acquisition of data; and KB, CM, SW, KH, RS, DS, MH-H, PB, IS and WG to data analysis and interpretation; KB, CM and PH drafted the article, and all authors revised it critically and approved the final version for publication.
Footnotes
The authors declare that they have no conflict of interest.
References
- Achard P, Cheng H, Grauwe LD, Decat J, Schoutteten H, Moritz T, Straeten DVD, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Aguirrezabal L, Bouchier-Combaud S, Radziejwoski A, Dauzat M, Cookson SJ, Granier C (2006) Plasticity to soil water deficit in Arabidopsis thaliana: dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ 29: 2216–2227 [DOI] [PubMed] [Google Scholar]
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Anastasiou E, Lenhard M (2007) Growing up to one’s standard. Curr Opin Plant Biol 10: 63–69 [DOI] [PubMed] [Google Scholar]
- Baerenfaller K, Hirsch-Hoffmann M, Svozil J, Hull R, Russenberger D, Bischof S, Lu Q, Gruissem W, Baginsky S (2011) pep2pro: a new tool for comprehensive proteome data analysis to reveal information about organ-specific proteomes in Arabidopsis thaliana. Integr Biol Camb 3: 225–237 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Hurkman WJ, Buchanan BB (2006) Thioredoxin target proteins in chloroplast thylakoid membranes. Antioxid Redox Signal 8: 1829–1834 [DOI] [PubMed] [Google Scholar]
- Barkan A (2011) Expression of plastid genes: organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkoulas M, Galinha C, Grigg SP, Tsiantis M (2007) From genes to shape: regulatory interactions in leaf development. Curr Opin Plant Biol 10: 660–666 [DOI] [PubMed] [Google Scholar]
- Beemster GTS, Veylder LD, Vercruysse S, West G, Rombaut D, Hummelen PV, Galichet A, Gruissem W, Inzé D, Vuylsteke M (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57: 289–300 [Google Scholar]
- Berkowitz O, Jost R, Pollmann S, Masle J (2008) Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell 20: 3430–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler LV, Cramer R (2011) Quantitative plant proteomics. Proteomics 11: 756–775 [DOI] [PubMed] [Google Scholar]
- Bognár LK, Hall A, Adám E, Thain SC, Nagy F, Millar AJ (1999) The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B. Proc Natl Acad Sci USA 96: 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth-stage based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, Zhang C, Morris K, Jenner C, Jackson S, Thomas B, Tabrett A, Legaie R, Moore JD, Wild DL, Ott S et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyton C, Nandha B, Johnson GN, Joliot P, Finazzi G (2006) Redox modulation of cyclic electron flow around photosystem I in C3 plants. Biochemistry 45: 13465–13475 [DOI] [PubMed] [Google Scholar]
- Brioudes F, Thierry AM, Chambrier P, Mollereau B, Bendahmane M (2010) Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc Natl Acad Sci USA 107: 16384–16389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, Hub AO Group WPW. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics 25: 288–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Temple B, Liang J, Guo J, Alonso JM, Ecker JR, Jones AM (2006) RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot 57: 2697–2708 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson SJ, Radziejwoski A, Granier C (2006) Cell and leaf size plasticity in Arabidopsis: what is the role of endoreduplication? Plant Cell Environ 29: 1273–1283(2006) [DOI] [PubMed] [Google Scholar]
- Cookson SJ, Van Lijsebettens M, Granier C (2005) Correlation between leaf growth variables suggest intrinsic and early controls of leaf size in Arabidopsis thaliana. Plant Cell Environ 28: 1355–1366 [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL (2008) Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz de Carvalho MH (2008) Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal Behav 3: 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C (2009) Global signatures of protein and mRNA expression levels. Mol Biosyst 5: 1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG (1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Fabre J, Dauzat M, Nègre V, Wuyts N, Tireau A, Gennari E, Neveu P, Tisné S, Massonnet C, Hummel I, Granier C (2011) PHENOPSIS DB: an information system for Arabidopsis thaliana phenotypic data in an environmental context. BMC Plant Biol 11: 77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff NV, Battisti DS, Beachy RN, Cooper PJM, Fischhoff DA, Hodges CN, Knauf VC, Lobell D, Mazur BJ, Molden D, Reynolds MP, Ronald PC, Rosegrant MW, Sanchez PA, Vonshak A, Zhu JK . (2010) Radically rethinking agriculture for the 21st century. Science 327: 833–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AJ (2005) The control of leaf development. New Phytol 166: 9–20 [DOI] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MFO, Yariv I, Dor C, Bassani M (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36: 629–642 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M (2004) A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Vanhaeren H, Inzé D (2012) Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci 6: 332–340 [DOI] [PubMed] [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, Thioux JJ, Rolland G, Bouchier- Combaud S, Lebaudy A, Muller B, Simonneau T, Tardieu F (2006) PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol 169: 623–635 [DOI] [PubMed] [Google Scholar]
- Granier C, Tardieu F (1998) Spatial and temporal analyses of expansion and cell cycle in sunflower leaves. A common pattern of development for all zones of a leaf and different leaves of a plant. Plant Physiol 116: 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang S, Valerius O, Hall H, Zeng Q, Li JF, Weston DJ, Ellis BE, Chen JG (2011) Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol 155: 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clockcontrolled genes. Proc Natl Acad Sci USA 99: 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb A, Krishnan A, Ambavaram MMR, Pereira A (2010) Molecular and physiological analysis of drought stress in Arabidopsis reveals early responses leading to acclimation in plant growth. Plant Physiol 154: 1254–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Tsiantis M (2009) A KNOX family TALE. Curr Opin Plant Biol 12: 593–598 [DOI] [PubMed] [Google Scholar]
- Hirsch-Hoffmann M, Gruissem W, Baerenfaller K (2012) pep2pro: the high-throughput proteomics data processing, analysis, and visualization tool. Front Plant Sci 3: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Quail PH (2003) Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol 133: 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel I, Pantin F, Sulpice R, Piques M, Rolland G, Dauzat M, Christophe A, Pervent M, Bouteillé M, Stitt M, Gibon Y, Muller B (2010) Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiol 154: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram GC, Waites R (2006) Keeping it together: co-ordinating plant growth. Curr Opin Plant Biol 9: 12–20 [DOI] [PubMed] [Google Scholar]
- Jansen R, Greenbaum D, Gerstein M (2002) Relating whole-genome expression data with protein-protein interactions. Genome Res 12: 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS (2010) Addressing accuracy and precision issues in iTRAQ quantitation. Mol Cell Proteomics 9: 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P, Gloor G (2001) The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6: 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Singh S, Wensel A, Huala E (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MV, Topper SE, Hubler SL, Hose J, Wenger CD, Coon JJ, Gasch AP (2011) A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol Syst Biol 7: 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs-Mache S (2011) Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription? Plant Mol Biol 76: 235–249 [DOI] [PubMed] [Google Scholar]
- Lidder P, Gutiérrez RA, Salomé PA, McClung CR, Green PJ (2005) Circadian control of messenger RNA stability. Association with a sequence-specific messenger RNA decay pathway. Plant Physiol 138: 2374–2385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K, Weihe A, Börner T (2011) The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J Plant Physiol 168: 1345–1360 [DOI] [PubMed] [Google Scholar]
- Locke JCW, Southern MM, Kozma-Bognár L, Hibberd V, Brown PE, Turner MS, Millar AJ (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol Syst Biol 1: 0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Rate DN, Song JT, Greenberg JT (2003) ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier T, Güell M, Serrano L (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett 583: 3966–3973 [DOI] [PubMed] [Google Scholar]
- Maier T, Schmidt A, Güell M, Kühner S, Gavin AC, Aebersold R, Serrano L (2011) Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol 7: 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MD, Wenger JP, Gilding E, Jilk R, Dixon RA (2009) Transcriptome analysis of Arabidopsis wildtype and gl3-sst sim trichomes identifies four additional genes required for trichome development. Mol Plant 2: 803–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet C, Tisné S, Radziejwoski A, Vile D, Veylder LD, Dauzat M, Granier C (2011) New insights into the control of endoreduplication: endoreduplication could be driven by organ growth in Arabidopsis leaves. Plant Physiol 157: 2044–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet C, Vile D, Fabre J, Hannah MA, Caldana C, Lisec J, Beemster GTS, Meyer RC, Messerli G, Gronlund JT, Perkovic J, Wigmore E, May S, Bevan MW, Meyer C, Rubio-Díaz S, Weigel D, Micol JL, Buchanan-Wollaston V, Fiorani F et al. (2010) Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiol 152: 2142–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo AK, Marder JB, Edelman M (1989) Dynamics of the photosystem II reaction center. Cell 56: 241–246 [DOI] [PubMed] [Google Scholar]
- McIntosh KB, Bonham-Smith PC (2006) Ribosomal protein gene regulation: what about plants? Canad J Bot Revue Canadienne De Botanique 84: 342–362 [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ 33: 453–467 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JDG (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Nozue K, Maloof JN (2006) Diurnal regulation of plant growth. Plant Cell Environ 29: 396–408 [DOI] [PubMed] [Google Scholar]
- Onda Y, Yagi Y, Saito Y, Takenaka N, Toyoshima Y (2008) Light induction of Arabidopsis SIG1 and SIG5 transcripts in mature leaves: differential roles of cryptochrome 1 and cryptochrome 2 and dual function of SIG5 in the recognition of plastid promoters. Plant J 55: 968–978 [DOI] [PubMed] [Google Scholar]
- Pantin F, Simonneau T, Rolland G, Dauzat M, Muller B (2011) Control of leaf expansion: a developmental switch from metabolics to hydraulics. Plant Physiol 156: 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A, Unwin RD, Evans CA, Griffiths S, Carney L, Zhang L, Jaworska E, Lee CF, Blinco D, Okoniewski MJ, Miller CJ, Bitton DA, Spooncer E, Whetton AD (2008) Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Mol Cell Proteomics 7: 853–63 [DOI] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Höhne M, Usadel B, Gibon Y, Rohwer J, Stitt M (2009) Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol Syst Biol 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar V, Löttgert T, Gigolashvili T, Bell K, Flügge UI, Häusler RE (2009) Molecular and functional characterization of the plastid-localized Phosphoenolpyruvate enolase (ENO1) from Arabidopsis thaliana. FEBS Lett 583: 983–991 [DOI] [PubMed] [Google Scholar]
- R Development Core Team. (2010) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, ISBN 3-900051-07-0 [Google Scholar]
- Refrégier G, Pelletier S, Jaillard D, Höfte H (2004) Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiol 135: 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehrauer H, Aquino C, Gruissem W, Henz SR, Hilson P, Laubinger S, Naouar N, Patrignani A, Rombauts S, Shu H, de Peer YV, Vuylsteke M, Weigel D, Zeller G, Hennig L (2010) AGRONOMICS1: a new resource for Arabidopsis transcriptome profiling. Plant Physiol 152: 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–1169 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18: 1292–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster TA, Queitsch C (2005) The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr Opin Plant Biol 8: 86–92 [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K (1996) Molecular responses to drought and cold stress. Curr Opin Biotechnol 7: 161–167 [DOI] [PubMed] [Google Scholar]
- Skirycz A, Bodt SD, Obata T, Clercq ID, Claeys H, Rycke RD, Andriankaja M, Aken OV, Breusegem FV, Fernie AR, Inzé D (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Inzé D (2010) More from less: plant growth under limited water. Curr Opin Biotechnol 21: 197–203 [DOI] [PubMed] [Google Scholar]
- Smith AM, Stitt M (2007) Coordination of carbon supply and plant growth. Plant Cell Environ 30: 1126–1149 [DOI] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, Vorwerk S, Youngs H (2004) Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Cosgrove DJ (2009) Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr Biol 19: R800–R811 [DOI] [PubMed] [Google Scholar]
- Tardieu F, Granier C, Muller B (2011) Water deficit and growth. Co-ordinating processes without an orchestrator? Curr Opin Plant Biol 14: 283–289 [DOI] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Tillich M, Hardel SL, Kupsch C, Armbruster U, Delannoy E, Gualberto JM, Lehwark P, Leister D, Small ID, Schmitz-Linneweber C (2009) Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc Natl Acad Sci USA 106: 6002–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisné S, Schmalenbach I, Reymond M, Dauzat M, Pervent M, Vile D, Granier C (2010) Keep on growing under drought: genetic and developmental bases of the response of rosette area using a recombinant inbred line population. Plant Cell Environ 33: 1875–1887 [DOI] [PubMed] [Google Scholar]
- Trémousaygue D, Garnier L, Bardet C, Dabos P, Hervé C, Lescure B (2003) Internal telomeric repeats and ’TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J 33: 957–966 [DOI] [PubMed] [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y (2009) Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol 150: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK (2006) Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 45: 523–539 [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Côté R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res 38: D736–D742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, de Sousa Abreu R, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO (2010) Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GY, Shi JL, Ng G, Battle SL, Zhang C, Lu H (2011) Circadian clock-regulated phosphate transporter PHT4;1 plays an important role in Arabidopsis defense. Mol Plant 4: 516–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts N, Massonnet C, Dauzat M, Granier C (2012) Structural assessment of the impact of environmental constraints on Arabidopsis thaliana leaf growth: a 3D approach. Plant Cell Environ 35: 1631–1646 [DOI] [PubMed] [Google Scholar]
- Wuyts N, Palauqui JC, Conejero G, Verdeil JL, Granier C, Massonnet C (2010) High-contrast three-dimensional imaging of the Arabidopsis leaf enables the analysis of cell dimensions in the epidermis and mesophyll. Plant Methods 6: 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky CM, Bickel DR (2010) Validation of differential gene expression algorithms: application comparing fold-change estimation to hypothesis testing. BMC Bioinformatics 11: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller G, Henz SR, Widmer CK, Sachsenberg T, Rätsch G, Weigel D, Laubinger S (2009) Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J 58: 1068–1082 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1-14, Supplementary tables S1-30