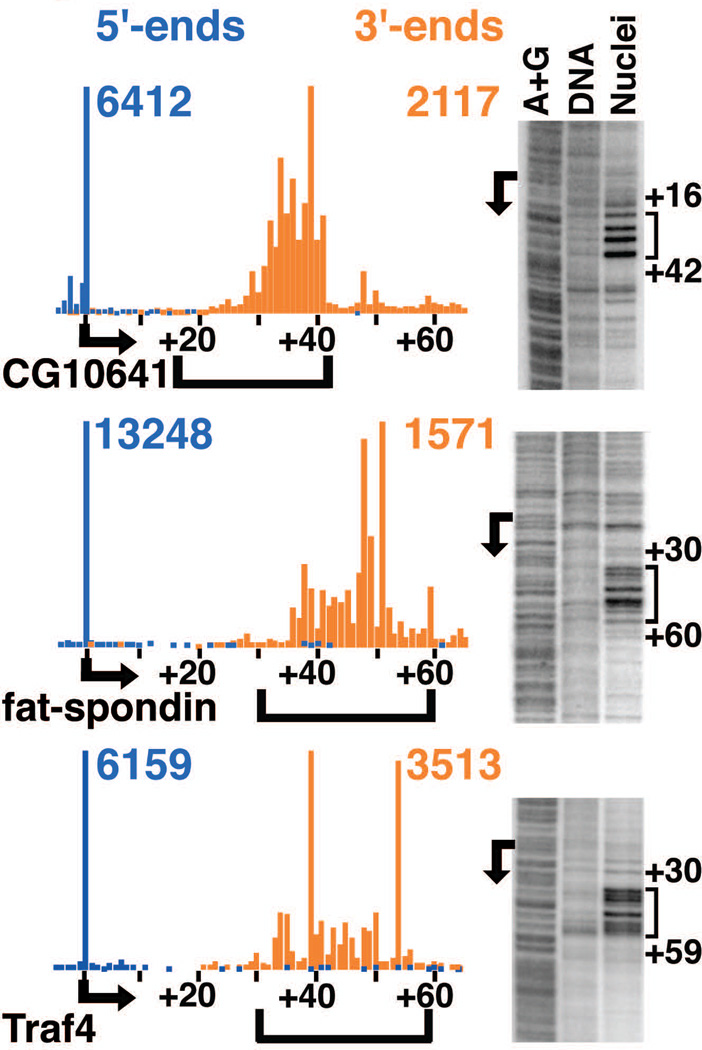
Fig. 2. 3’-ends of short RNAs identify sites of polymerase stalling.
5’-end (blue) and 3’-end (orange) RNA reads are shown for example genes, designated by Flybase gene names, with observed TSSs indicated by arrows. The number of 5’- and 3’-end reads at the peak locations is given in the corresponding colors. Permanganate footprints from nuclei are shown for each gene, alongside purified DNA and the A+G DNA ladder. Numbers in black indicate positions relative to the TSS, and brackets show areas of permanganate reactivity.