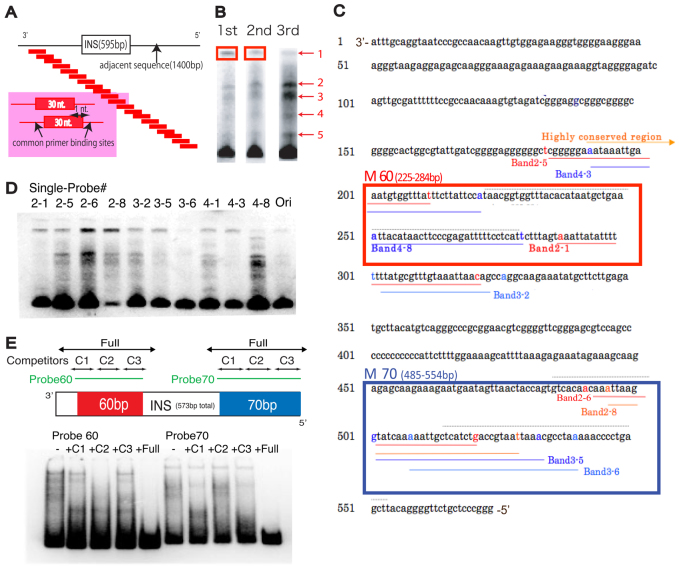
Fig. 1.
The mega-shift assay identifies regions of protein binding. (A) Experimental design of the mega-shift assay. A tiled pool of 30-mer probes, each shifted by one nucleotide from the adjacent probe and containing common primer sites, were designed to cover 2 kb of total sequence flanking and including the ArsI site. (B) Probes extracted from the top band (boxed) were amplified and used as probes for the next round of mega-shift assay. Three rounds of mega-shift resulted in enriched pools of primers with five different but consistent complexes. Probes were extracted from each of the five bands and sequenced. (C) Probes sequenced from each band obtained in B were mapped to the ArsI element. Probes derived from diverse complexes were found to be overlapping in two regions of the ArsI sequence: M-60 and M-70. Each band number corresponds to the single probe #. (D) A single probe designed for each region labeled in C was used for a gel-shift assay and resulted in several bands, suggesting that ArsI-associated proteins might form a higher-order protein complex. (E) Competitors designed for each region (C1-C3) and the entire region (Full) within M-60 and M-70 of ArsI did not compete as effectively as the full-length competitor, indicating the existence of multiple protein binding sites.