Fig. 1.
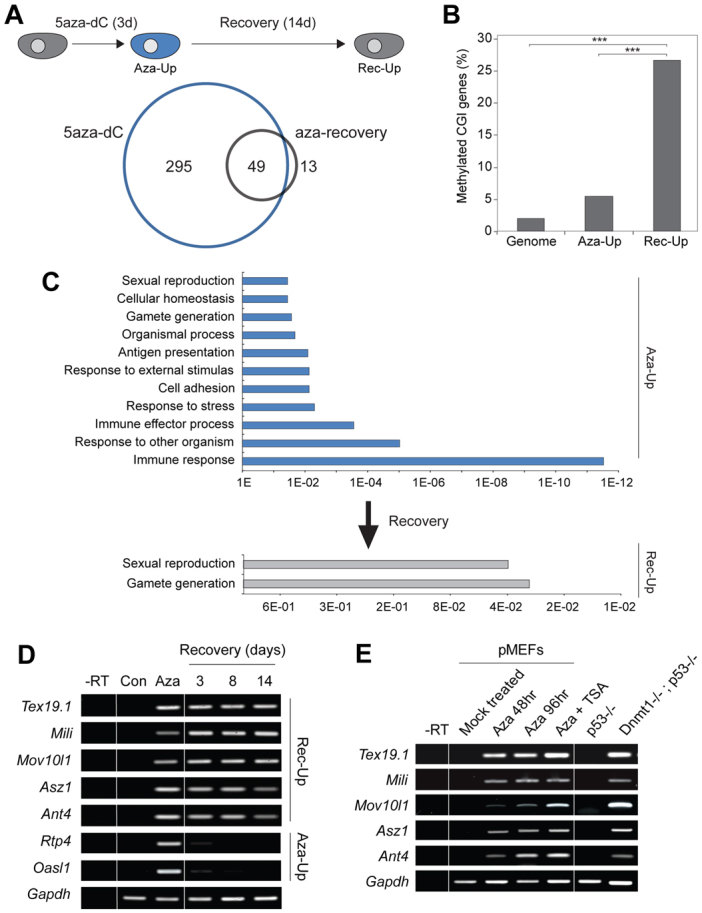
Identification of core methylation-dependent genes. (A) Schematic of the 5aza-dC-recovery assay. Cells are exposed to 5aza-dC for 3 days (3d) and allowed to recover for 14 days (14d). Below: Venn diagram showing overlap between genes upregulated after 5aza-dC (Aza-Up; blue) and after recovery (Rec-Up; grey). (B) Percentage of genes with somatically methylated CpG island (CGI) promoters in Aza-Up or after 14 days recovery (Rec-Up) (Meissner et al., 2008). ***P<0.001. (C) Gene ontology of Aza-Up and Rec-Up gene set, with P-values along the x-axis. Multiple categories of genes are activated by 5aza-dC exposure but only germline-associated categories remain enriched after recovery, when only methylation-dependent genes are predicted to remain. (D) RT-PCR validation of selected germline Rec-Up genes from microarray analysis that remain active during recovery and of control somatic Aza-Up genes that re-impose silencing after 5aza-dC exposure, probably owing to indirect activation. Gapdh is a loading control. (E) RT-PCR showing that genome-defence genes are activated by demethylation in multiple contexts including in Dnmt1n/n MEFs and by 5aza-dC (± the histone deacetylase inhibitor TSA) in primary MEFs. –RT, without reverse transcriptase.
