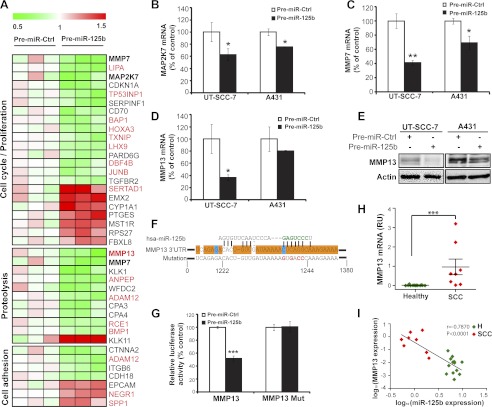
FIGURE 5.
Identification of genes regulated by miR-125b in cSCC cells. A, microarray analysis performed in independent biological triplicates for UT-SCC-7 cells transfected with either Pre-miR-125b or Pre-miR-Ctrl. Heat map showing the relative expression of miR-125b regulated genes involved in proliferation, proteolysis and cell adhesion. Putative miR-125b targets predicted by miRWalk are labeled in red. B, miR-125b mediated regulation for MAP2K7 (B), MMP7 (C), and MMP13 (D) was verified in both cSCC cell lines by qRT-PCR. *, p < 0.05, **, p < 0.01. Student's t test. E, cSCC cells were transfected with Pre-miR-125b or Pre-miR-Ctrl for 48 h and MMP13 protein was detected by Western blotting. β-actin was detected on the same blot as loading control. F, nucleotide resolution of the predicted miR-125b target site in 3′-UTR of MMP13 mRNA: the seed sequence (green letters); the evolutionarily conserved regions in at least 12 species (orange boxes) or in 6 species (blue boxes); the mutant miR-125b binding sites (red letters). G, UT-SCC-7 cells were transfected with luciferase reporter construct containing MMP13 3′UTR together with Pre-miR-125b or Pre-miR-Ctrl. ***, p < 0.001. Student's t test. H, expression of MMP13 mRNA was analyzed in healthy skin (n = 13) and cSCC biopsies (n = 8) using qRT-PCR. ***, p < 0.001, Mann-Whitney test. I, correlation of miR-125b with MMP13 expression in cSCC and healthy skin. Spearman-Correlation on log-transformed values.