Background: WT non-visual arrestins are promiscuous, binding numerous GPCRs.
Results: Mutations of very few receptor discriminator residues greatly increase receptor specificity of arrestin-3.
Conclusion: Targeted manipulation of key residues that determine receptor preference is a viable approach to the construction of arrestins with high specificity for particular GPCR subtypes.
Significance: Non-visual arrestins with high receptor specificity make therapeutic use of signaling-biased arrestin mutants feasible.
Keywords: 7-Helix Receptor, Adrenergic Receptor, Arrestin, Dopamine Receptors, Protein Engineering, Signal Transduction, Rhodopsin
Abstract
Based on the identification of residues that determine receptor selectivity of arrestins and the analysis of the evolution in the arrestin family, we introduced 10 mutations of “receptor discriminator” residues in arrestin-3. The recruitment of these mutants to M2 muscarinic (M2R), D1 (D1R) and D2 (D2R) dopamine, and β2-adrenergic receptors (β2AR) was assessed using bioluminescence resonance energy transfer-based assays in cells. Seven of 10 mutations differentially affected arrestin-3 binding to individual receptors. D260K and Q262P reduced the binding to β2AR, much more than to other receptors. The combination D260K/Q262P virtually eliminated β2AR binding while preserving the interactions with M2R, D1R, and D2R. Conversely, Y239T enhanced arrestin-3 binding to β2AR and reduced the binding to M2R, D1R, and D2R, whereas Q256Y selectively reduced recruitment to D2R. The Y239T/Q256Y combination virtually eliminated the binding to D2R and reduced the binding to β2AR and M2R, yielding a mutant with high selectivity for D1R. Eleven of 12 mutations significantly changed the binding to light-activated phosphorhodopsin. Thus, manipulation of key residues on the receptor-binding surface modifies receptor preference, enabling the construction of non-visual arrestins specific for particular receptor subtypes. These findings pave the way to the construction of signaling-biased arrestins targeting the receptor of choice for research or therapeutic purposes.
Introduction
G protein-coupled receptors (GPCRs)2 are the largest and the most functionally and structurally diverse family of signaling proteins in mammals (1, 2). Different species have from 800 to >3,400 GPCR subtypes encoded by 3–10% of their genes (SEVENS database, available on the Computational Biology Research Center Web site). Different GPCRs respond to a wide variety of stimuli, from light, small molecules, and extracellular calcium to peptide and protein hormones and extracellular protease activity. Active receptors sequentially activate multiple G protein molecules, amplifying the signal. This process is stopped when G protein-coupled receptor kinases selectively phosphorylate activated receptors (3) and arrestins bind to active phosphoreceptor (4), blocking further G protein coupling by steric exclusion (5, 6). Mammals have only seven G protein-coupled receptor kinases that serve hundreds of different GPCRs (7, 8), whereas the complement of arrestins is even smaller, only four subtypes (9). Arrestin-13 and -4 are specifically expressed in photoreceptor cells and quench light-induced signaling by rhodopsin and cone opsins (10, 11). In contrast, arrestin-2 and -3 are expressed in virtually every cell in the body and regulate the great majority of GPCRs (10). In most cells, including mature neurons that express the highest levels of non-visual arrestins, arrestin-2 outnumbers arrestin-3 by ∼10–20:1 (12, 13). Thus, evolution produced arrestins with high receptor specificity, such as arrestin-1 with high preference for rhodopsin (14–16), along with fairly promiscuous non-visual arrestins (16, 17).
Several genetic disorders are associated with mutations in GPCRs (18). In the case of loss-of-function mutations, gene replacement therapy introducing a functional version of the affected protein is the most logical therapeutic approach. However, the treatment of patients with gain-of-function mutations requires a different strategy; these mutations are dominant, which means that the other perfectly normal allele does not help. Excessive GPCR activity can be dampened by enhanced arrestins with preactivating mutations in vitro (19), in Xenopus oocytes (20–22), and even in living animals (23). This compensational approach has been recently shown to work in the visual system; phosphorylation-independent arrestin-1 expressed in rods with defective rhodopsin phosphorylation was shown to improve overall health of photoreceptors and their functional performance (23). Homologous mutations in all arrestins yield enhanced forms that bind phosphorylated and especially active unphosphorylated receptors much better than wild type (WT) arrestins (20, 22, 24, 25). Whereas in rods, a single receptor, rhodopsin, along with its cognate arrestin-1 clearly predominate, most cells express a dozen or more GPCR subtypes and two non-visual arrestins with low specificity. Thus, the introduction into the cell of the existing enhanced mutants of arrestin-2 or -3 would reduce excessive signaling by mutant receptors while at the same time attenuating perfectly normal signaling by other GPCRs. This problem can be solved by the construction of non-visual arrestins with dramatically increased preference for groups of GPCRs or individual subtypes.
Arrestin elements that determine receptor preference are localized on the concave side of both N- and C-domain of arrestin proteins (15). Recent identification of only 10 residues that largely define receptor specificity and drive arrestin-GPCR interactions (16) sets the stage for the construction of mutants with significantly enhanced receptor specificity. Here we show that targeted manipulation of a few receptor discriminator residues differentially changes the ability of arrestin-3 to bind the M2 muscarinic (M2R), β2-adrenergic (β2AR), and D1 (D1R) and D2 (D2R) dopamine receptors, thereby shifting its preference among individual GPCRs.
EXPERIMENTAL PROCEDURES
Materials
[γ-32P]ATP, [14C]leucine, and [3H]leucine were from PerkinElmer Life Sciences. All restriction and DNA-modifying enzymes were from New England Biolabs (Ipswich, MA). Rabbit reticulocyte lysate was from Ambion (Austin, TX), and SP6 RNA polymerase was prepared as described (26). Cell culture reagents and media were from Mediatech (Manassas, VA) or Invitrogen. The luciferase substrate coelenterazine-h was from DiscoveRx (Fremont, CA). All other reagents were from Amresco (Solon, OH) or Sigma-Aldrich.
Mutagenesis and Plasmid Construction
Plasmids that encode the prevalent short splice variant of arrestin-3 (27, 28) with engineered unique restriction sites and arrestin-3-NCA mutant with all key receptor-binding residues replaced with alanines were described previously (29, 30). All mutations were introduced by PCR, using the strategy recently described (30). All constructs were confirmed by dideoxy sequencing. All arrestin mutants were N-terminally tagged with Venus, whereas the receptors were C-terminally tagged with Renilla luciferase variant 8 (RLuc8), as described (16, 30, 31). Untagged arrestins were also subcloned into pGEM-2 in vitro transcription plasmid (Promega, Madison, WI).
In Vitro Transcription, Translation, Rhodopsin Preparation, and Light-activated Phosphorylated Rhodopsin (P-Rh*) Binding Assay
These were performed as described recently (16).
Bioluminescence Resonance Energy Transfer (BRET) Assay
BRET-based arrestin-receptor interaction assays (32, 33) were performed and analyzed, as described (30). The appropriate agonists (25 μm carbamylcholine for M2R, 10 μm isoproterenol for β2AR, 10 μm dopamine for D1R, or 10 μm quinpirole for D2R) were added at 37 °C, 15 min prior to the addition of 5 μm luciferase substrate coelenterazine-h.
Receptor Activity Assays
cAMP in live cells was measured using GloSensor (clone 22f) (Promega), as described in the legend to supplemental Fig. S1. GloSensor 22f is a genetically modified form of firefly luciferase with inserted cAMP-binding protein moiety. Binding of cAMP induces a conformational change, leading to increased light output (34–36). Receptor-fused Renilla luciferase cannot use the substrate of firefly luciferase and therefore does not generate interfering background luminescence. The results indicated that Renilla luciferase-tagged receptors were functional (supplemental Fig. S1). In all cases, tagged receptors showed lower ability to couple to their cognate G proteins than WT controls, in line with an earlier report that β2AR-GFP (which is similar in size to luciferase) is active but less potent than untagged β2AR (37).
Data Analysis and Statistics
BRET and direct binding assay data were analyzed, as described (16, 30). Statistical significance was determined by one-way ANOVA with post hoc Dunnett's test with correction for multiple comparisons, or two-way ANOVA followed by a Holm-Sidak test to compare the effect of mutations and receptor type on BRET. GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA) was used for curve fitting and statistical analysis.
RESULTS
Selection of Parental Arrestin for the Construction of Receptor-specific Mutants
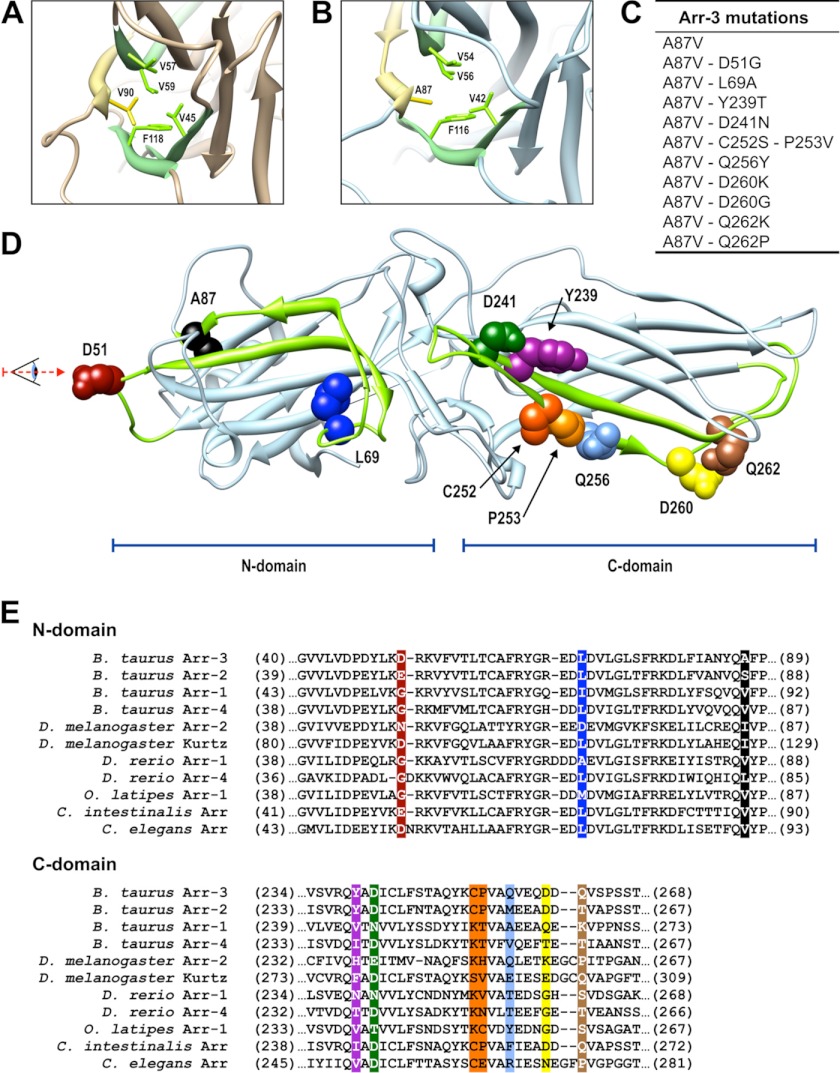
Both non-visual arrestins are fairly promiscuous, capable of interacting with many of the GPCRs tested so far (14, 17, 20, 22, 25, 38, 39). However, arrestin-3 appears to have a wider receptor repertoire and binds several GPCRs with higher affinity than arrestin-2 (17, 40, 41). Therefore, we chose arrestin-3 as the parental subtype for the generation of the mutants used in this study. Arrestin-1 is the only subtype that demonstrates high receptor specificity, preferentially binding to light-activated phosphorhodopsin (14, 16, 30). The comparison of the crystal structures of arrestin-1 (42) with those of non-visual arrestin-2 (43, 44) and -3 (45) identified one feature directly related to receptor specificity. In arrestin-1, Val-90 is localized between the two sheets of the β-strand sandwich of the N-domain. This residue participates in multiple interactions with other bulky hydrophobic residues (Val-45, Val-57, Val-59, and Phe-118), apparently stabilizing the N-domain (42) (Fig. 1A). This valine is conspicuously absent in both non-visual subtypes, where it is replaced with serine (Ser-86 in arrestin-2 (43)) or alanine (Ala-87 in arrestin-3 (45)). Interestingly, all of its potential interaction partners are conserved in arrestin-3 (Val-42, Val-54, Val-56, and Phe-116) (Fig. 1B). It appears that the absence of this valine in non-visual arrestins makes the N-domain more flexible. Arrestin-1 shows very little binding to the phosphorylated active M2R, whereas arrestin-2 binds this receptor well (14, 15). The V90S mutation in arrestin-1, mimicking the situation in arrestin-2, dramatically reduces the receptor specificity of arrestin-1, enhancing its binding to M2R virtually to the level shown by arrestin-2 (43). These data suggest that to increase receptor specificity of arrestin-3, it is necessary to make its N-domain more rigid, mimicking that of arrestin-1, so that it would be harder for the resulting mutant to “mold” itself successfully on every GPCR it encounters. Therefore, we introduced the A87V mutation into arrestin-3 and compared the binding of this mutant to M2R, β2AR, D1R, and D2R. Similar to the previous report that the S86V mutation in arrestin-2 does not appreciably affect its binding to M2R (43), we found that the A87V substitution in arrestin-3 does not significantly change its binding to any of the GPCRs tested (Fig. 2). Therefore, we used arrestin-3-A87V as the base mutant, which is likely to have a predisposition for higher receptor specificity due to a more rigid N-domain while retaining the ability to bind multiple GPCRs.
FIGURE 1.
Arrestin residues targeted in this study. A and B, crystal structure of arrestin-1 (Protein Data Bank entry 1CF1) (A) and arrestin-3 (Protein Data Bank entry 3P2D) (B), focusing on the core of the N-domain (viewed from the direction indicated in D). Side chains of Val-90 and Ala-87 (in arrestin-1 and -3, respectively) are shown in yellow, and adjacent hydrophobic residues are shown in green. C, arrestin-3 mutations introduced in this study. D, the receptor-binding surface of arrestin-3. Elements responsible for receptor preference are shown in green (β-strands V and IV in the N-domain and β-strands XV and XVI in the C-domain). The residues targeted in this study are shown as CPK models using the same color scheme as in E and in subsequent figures. E, sequence alignment of “receptor discriminator” elements of arrestins from different species (based on Ref. 9). Note that some of the homologous residues from other arrestins were introduced into arrestin-3 (C).
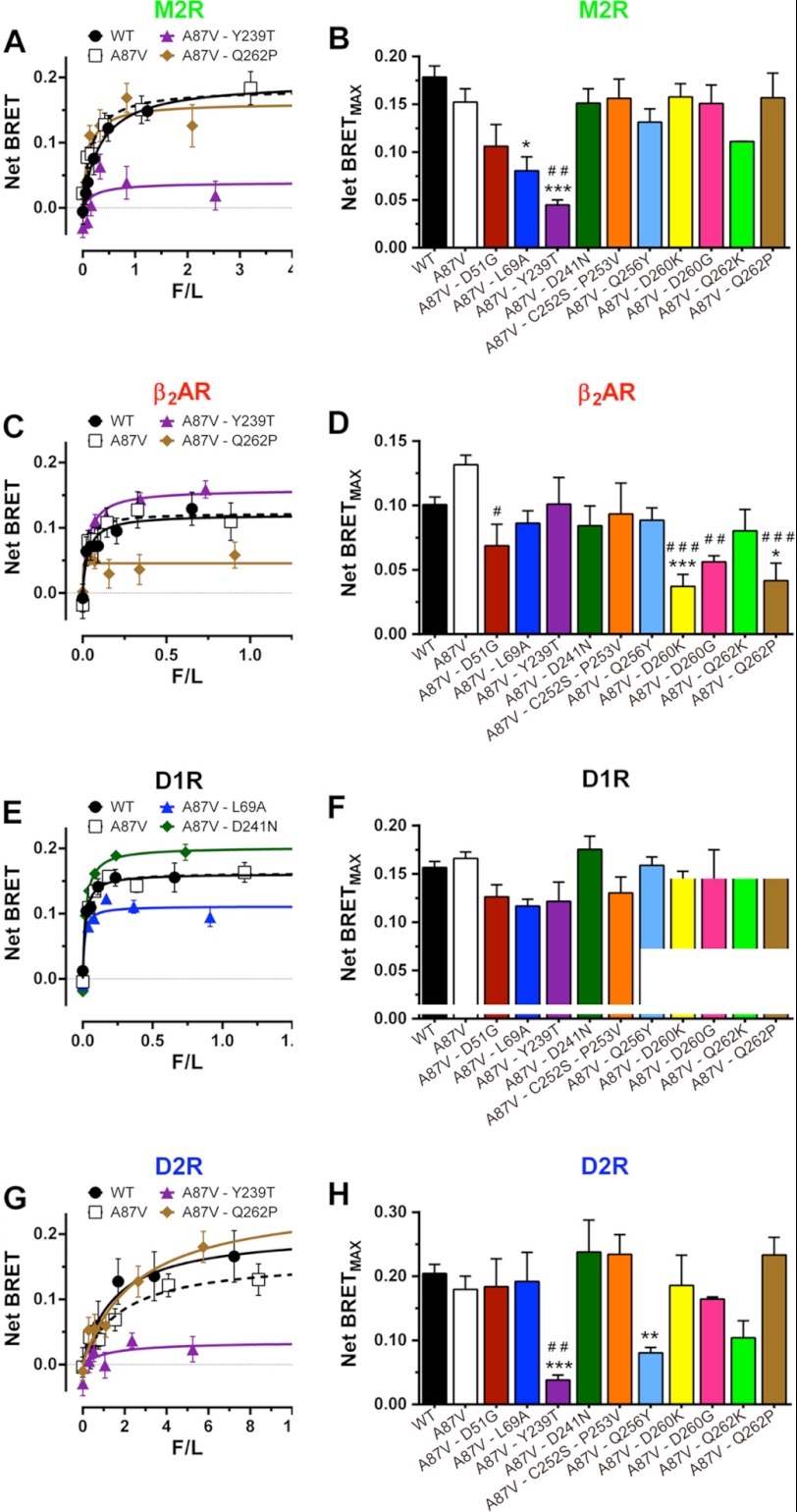
FIGURE 2.
Substitutions of receptor discriminator residues in arrestin-3 differentially affect agonist-induced increase in binding to individual GPCRs. BRET between indicated Venus-tagged arrestins and luciferase-tagged human M2R (A and B), β2AR (C and D), D1R (E and F), or D2R (G and H) in COS-7 cells. A, C, E, and G, net BRET (agonist-induced increase in BRET signal) as a function of Venus-arrestin expression level (measured by fluorescence) normalized by receptor level (luciferase luminescence) (F/L) (for details, see supplemental Fig. S2). Shown are means ± S.E. (error bars) of six repeats in a representative experiment (of 3–15 performed) for WT arrestin-3, A87V base mutant, and selected mutants with high or low net BRET. B, D, F, and H, net BRETMAX for the indicated mutant-receptor combinations (raw data shown in supplemental Figs. S3–S6). BRETMAX ± S.E. averaged across experiments is shown. Statistical significance was determined using one-way ANOVA with Dunnett's multiple comparison test. *, p < 0.05; **, p < 0.01; ***, p < 0.001, as compared with WT arrestin-3. #, p < 0.05; ##, p < 0.01; ###, p < 0.001, as compared with A87V base mutant.
Substitutions of Key Receptor Discriminator Residues Differentially Affect Arrestin Binding to Individual GPCRs
Arrestins are elongated two-domain molecules (Fig. 1D). An extensive surface encompassing the concave sides of both domains was implicated in receptor binding by a variety of methods (46–48). On this surface, we have previously identified four and six residues in the N- and C-domain, respectively, that are largely responsible for receptor preference (16). Alanine substitution of all of these residues in arrestin-1, -2, and -3 yields mutants essentially lacking the ability to bind receptors (16, 30), suggesting that these elements largely drive the interaction. Thus, these 10 residues are the most logical targets for manipulation of receptor specificity (Fig. 1, C and E). If any of all 20 possible residues could occupy each of these positions, the number of combinations would be enormous (2010). However, sequence comparison of cloned arrestins shows that relatively few residues could be found in each of these positions throughout >600 million years of arrestin evolution (9). Therefore, we substituted each residue only with those that are found in the corresponding position in other arrestin proteins (Fig. 1E).
Homologues of Asp-51 in arrestin-3 include Glu, Asn, Gln, and Gly. Because Gly is the least conservative substitution, we chose the D51G mutation. Homologues of Leu-69 include Ile, Met, His, Ala, and Asp, so we chose one of the non-conservative substitutions, L69A. Homologues of Tyr-239 are Leu, Ile, Val, Phe, Asn, His, Gly, and Thr, and we chose the latter. Asp-241 is less variable, with Glu, Thr, and Asn found in homologous positions. The pair Cys-252/Pro-253 is conserved in all vertebrate non-visual subtypes, but in other arrestins, Lys, Asn, or Ser replaces Cys, whereas Ile, Val, Asn, Thr, His, or Glu replaces Pro. Homologues of Gln-256 include hydrophobic residues and those with hydrogen-bonding capability, so we chose Tyr, which combines both features. Homologues of Asp-260 are quite variable, and insect and Caenorhabditis elegans arrestins have a two-residue insert after this position, so we chose two extremes, with charge reversal (D260K) and complete elimination of the side chain (D260G). Similarly, due to the high variability of Gln-262 homologues, we substituted it with the positively charged lysine or proline, which breaks secondary structure. Mutants with these 10 substitutions were generated on the background of arrestin-3-A87V (Fig. 1C), and their binding to M2R, β2AR, D1R, and D2R was tested in a cell-based assay, using BRET between luciferase-tagged receptors and Venus-tagged arrestins (Fig. 2 and supplemental Figs. S3–S6). In all cases, we used receptor binding-deficient arrestin-3-NCA (30) to evaluate the nonspecific “bystander” BRET (supplemental Figs. S2–S7).
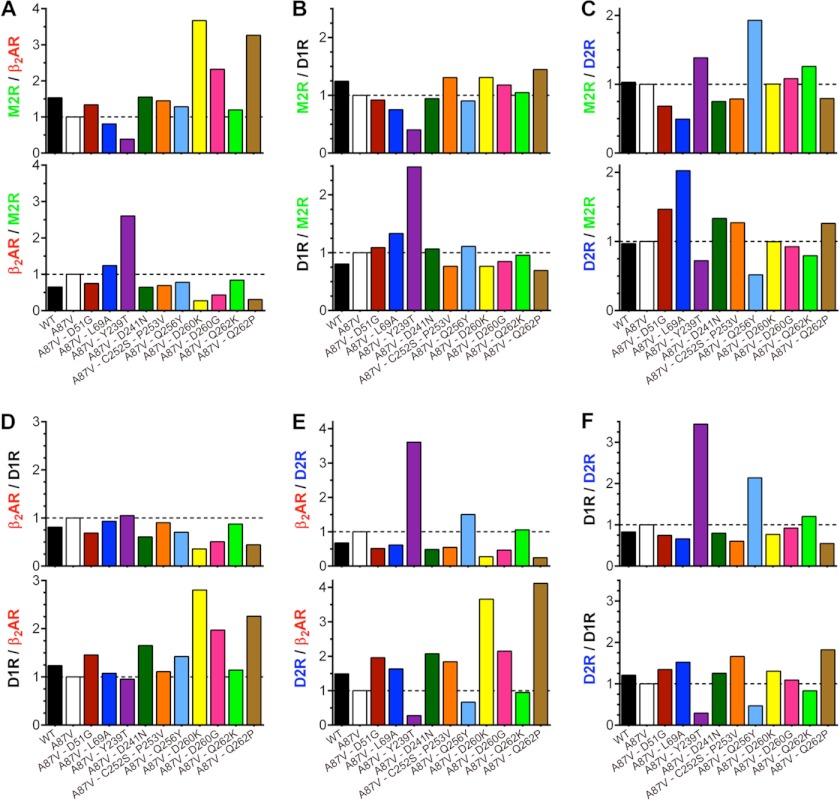
Only three of these mutations did not significantly affect arrestin binding to any of these four GPCRs: D241N, C252S/P253V, and Q262K (the latter showed a tendency to reduce the interaction with all four receptors, but the differences did not reach statistical significance). Six mutations significantly reduced the binding to just one of the four receptors; L69A reduced binding to M2R, and D51G, D260K, D260G, and Q262P reduced binding to β2AR, whereas Y239T reduced binding to M2R and D2R but not to β2AR or D1R (Fig. 2). The magnitude of these reductions varied from 2- to ∼5-fold. Interestingly, none of the mutations significantly increased arrestin binding, except D241N, which showed a tendency of doing so in the case of both dopamine receptors (Fig. 2). Considering how many arrestin residues participate in receptor binding (16, 46, 47), the magnitude of the changes due to point mutations is quite impressive. Most importantly, virtually all effects are receptor-specific (i.e. the mutations reduce the binding to a particular receptor but not to others). In order to gauge how much each mutation affected receptor preference, we calculated the selectivity index as the relative binding within receptor pairs, setting the binding ratio of the arrestin-3-A87V mutant at 1 (Fig. 3). The data show that single mutations change the relative binding to different GPCRs 2–4-fold.
FIGURE 3.
Receptor selectivity of arrestin-3 mutants. A–F, ratios of net BRETMAX (shown in Fig. 2) for the indicated mutants and receptor pairs are shown. For normalization, the binding ratio of the A87V base mutant was set at 1.
The Effects of Individual Mutation Are Additive
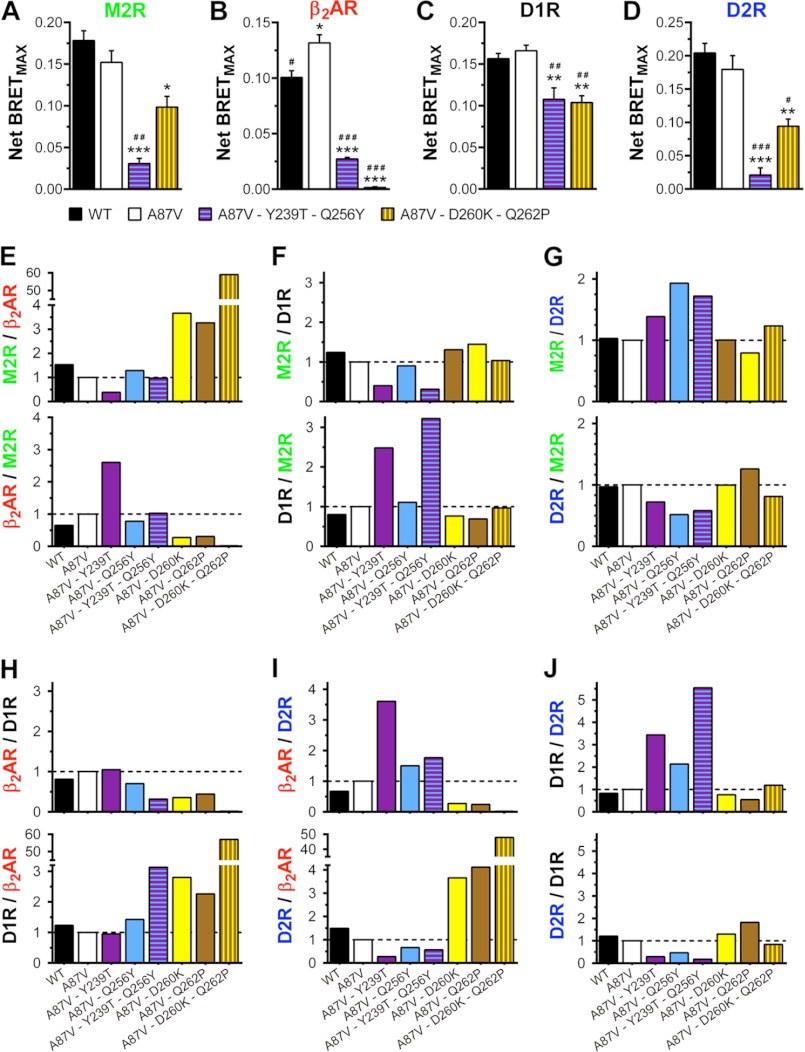
Certain point mutations on the receptor-binding surface of arrestin-3 change receptor preference in the same direction (Figs. 2 and 3). Both T239T and Q256Y significantly reduce arrestin binding to D2R, whereas D260K and Q262P suppress the interaction with β2AR (Figs. 2 and 3). To test whether we can further enhance receptor specificity by combining these substitutions, we constructed two double mutants, Y239T/Q256Y and D260K/Q262P, and tested their binding to the same set of receptors (Fig. 4). In contrast to the point mutations, each of these combinations appreciably changed the interactions with every GPCR (Fig. 4). However, the magnitude of the effects was very much receptor-dependent. Y239T/Q256Y significantly reduced the binding to M2R, β2AR, and D2R, with much smaller effect on D1R. D260K/Q262P only slightly reduced the interaction with M2R and D1R and decreased the binding to D2R by ∼50% while almost completely obliterating the ability of arrestin-3 to bind β2AR (Fig. 4, A–D). Thus, in the latter case, both the magnitude of the effect and its receptor selectivity are enhanced by the combination of two mutations, as compared with the parental single mutants (Fig. 4, E–J). Interestingly, although individual Y239T and Q256Y mutations only moderately reduce β2AR binding, their combination again shows an additive effect, significantly inhibiting arrestin-3 interactions with this receptor (Fig. 4J). Importantly, D260K/Q262P shows ∼50–60-fold preference for M2R, D1R, and D2R over β2AR (Fig. 4, E, H, and I), whereas Y239T/Q256Y shows ∼5-fold preference for D1R over D2R (Fig. 4J). These results show that the substitution of multiple residues on the receptor-binding surface of arrestin further enhances its receptor selectivity.
FIGURE 4.
Combinations Y239T/Q256Y and D260K/Q262P increase the effect of individual mutations on receptor preference. A–D, net BRETMAX (means ± S.E. (error bars) from 3–15 experiments performed with six repeats each) for the indicated arrestin-receptor combinations. Statistical significance of the differences, determined using one-way ANOVA with Dunnett's multiple comparison test, is indicated as follows. *, p < 0.05; **, p < 0.01; ***, p < 0.001, as compared with WT arrestin-3. #, p < 0.05; ##, p < 0.01; ###, p < 0.001, as compared with A87V base mutant. E–J, ratios of net BRETMAX (shown in A–D) for the indicated mutants and receptor pairs are shown. For normalization, the binding ratio of the A87V base mutant was set at 1.
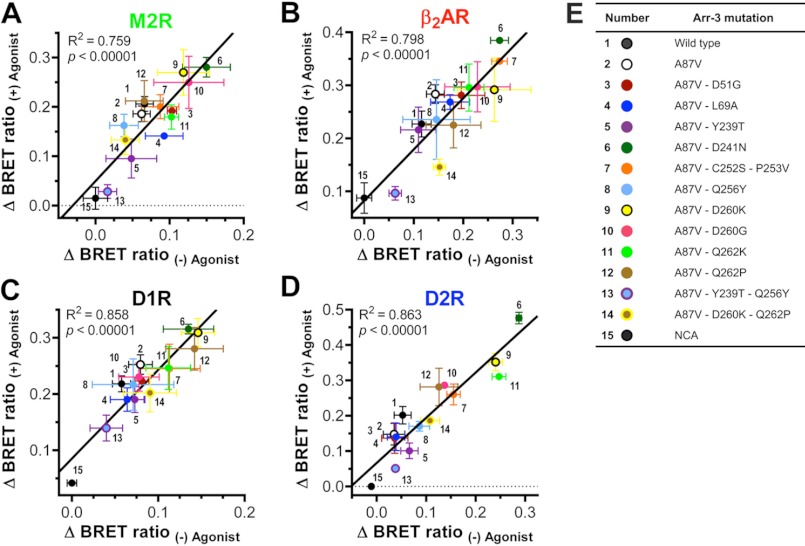
The Same Arrestin Elements Mediate Agonist-dependent and -independent Binding to All GPCRs
Previously, we found that arrestin interaction with receptors detected in intact cells using BRET between luciferase-tagged GPCRs and Venus-arrestins involves two components. Using free Venus and receptor binding-deficient Venus-arrestin-3-NCA mutant as negative controls, we detected measurable specific arrestin binding to M2R, β2AR, and D2R in the absence of agonists, which was considerably increased upon agonist stimulation (30). Basal interaction in the absence of agonists might reflect the level of constitutive activity of these receptors and/or the probability of arrestin “predocking” to inactive GPCRs, possibly enhanced by relatively high expression of Venus-arrestin at saturation of BRET signal. This phenomenon is reminiscent of “precoupling” of various GPCRs to their cognate G proteins that was reported in many cases (49–53). Regardless of the exact mechanism, it is important to establish whether both components of the arrestin-receptor interaction are mediated by the same arrestin elements. Therefore, for WT arrestin-3, its A87V base mutant, and all 12 mutants with altered receptor specificity, we determined the level of BRET signal obtained in the absence and presence of appropriate agonist for each of the four GPCRs. We found that upon subtraction of the signal obtained with the NCA mutant (reflecting nonspecific “bystander” BRET (30)), each mutation changed the levels of basal interaction and its agonist-induced increase in the same direction and to a similar extent (supplemental Fig. S7). Rigorous analysis of the data obtained with M2R, β2AR, D1R, and D2R shows that the two components of the interaction of the 15 arrestin-3 proteins with each receptor correlate very well (Fig. 5). Interestingly, some mutants show much higher basal binding and agonist-dependent increase than others. For example, D241N, D260K, and, to a lesser extent, D260G are always at the upper end of the curve, whereas Y239T and the same mutation combined with Q256Y tend to be closer to the binding-deficient NCA (Fig. 5). In cases like D241N, this propensity to bind appears to apply to all four GPCRs tested. Collectively, these results demonstrate that the same residues on the receptor-binding side of the two arrestin domains mediate both basal predocking and agonist-induced increase in arrestin binding to these GPCRs.
FIGURE 5.
Mutations in arrestin-3 similarly affect basal interactions and agonist-induced increases in binding to individual GPCRs. BRET signals obtained in the absence and presence of appropriate agonist of each receptor are shown in supplemental Figs. S3–S6. ΔBRET ratios were determined by subtracting the BRET ratio obtained with the receptor binding-deficient NCA mutant in the absence of agonist (nonspecific “bystander” BRET; supplemental Fig. S7). A–D, the correlation between ΔBRET ratios of the indicated mutants in the presence and absence of agonist for M2R (A), β2AR (B), D1R (C), and D2R (D). Goodness of fit for linear regression is reflected by the shown R2 and p values in each panel. High correlation shows that the same arrestin residues mediate both basal predocking and agonist-induced increase in arrestin binding to each GPCR. E, color scheme and number key. Error bars, S.E.
Mutations of Receptor Discriminator Residues Differentially Affect Arrestin-3 Binding to Rhodopsin
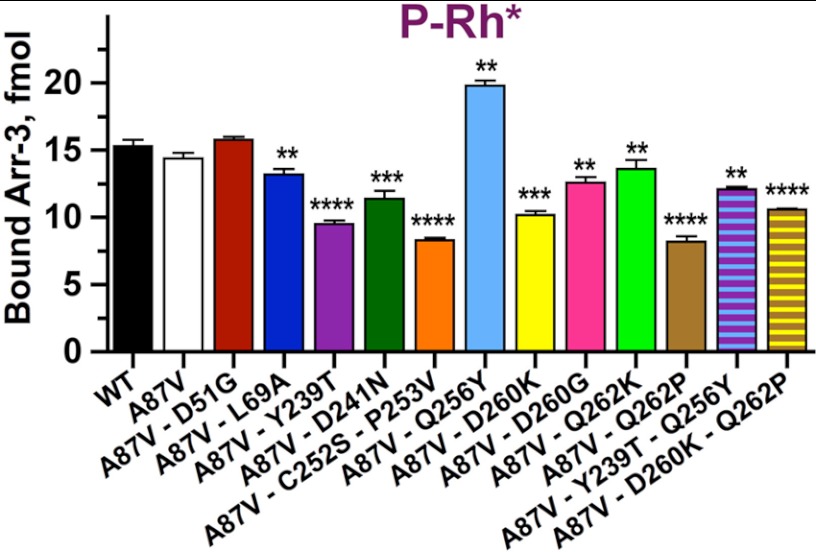
Originally, the 10 exposed side chains manipulated here were discovered as key determinants of arrestin-1 preference for rhodopsin and arrestin-2 specificity for non-visual GPCRs (16). Therefore, to further characterize mutant arrestins using an alternative method that does not involve large tags, BRET, or cultured cells, we used the well established direct in vitro binding assay with P-Rh* (16, 30, 54, 55). Untagged forms of all mutants were subcloned into a transcription vector, and radiolabeled arrestins were generated by cell-free translation. Comparison of all mutants with WT arrestin-3 showed that the A87V base mutation or its combination with D51G does not appreciably change P-Rh* binding, whereas the remaining 11 mutations demonstrate significant effects. Interestingly, Q256Y significantly increased P-Rh* binding of arrestin-3, whereas the rest of the mutations decreased it to different levels, some virtually by half (Fig. 6). These data independently confirmed that selected residues play a key role in receptor preference and that the results obtained in cells were not significantly affected by luciferase and Venus tags on receptors and arrestins, respectively, or by the method used to measure the arrestin-receptor interaction.
FIGURE 6.
Rhodopsin binding is differentially affected by mutations on the receptor-binding surface of arrestin-3. Translated radiolabeled WT arrestin-3 (WT) and the indicated mutants (50 fmol) were incubated with 0.3 μg of P-Rh* in 50 μl at room light at 37 °C for 5 min. The samples were cooled on ice, and bound arrestins were separated from free by gel filtration on 2-ml Sepharose 2B-CL columns, as described (74). Bound arrestins eluted with rhodopsin-containing membranes were quantified by scintillation counting. Means ± S.D. (error bars) of two independent experiments performed in duplicate are shown. The data were analyzed using one-way ANOVA with arrestin type as the main factor, followed by Dunnett's post hoc test with correction for multiple comparisons. Statistical significance of the differences is indicated as follows. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
DISCUSSION
Arrestins are elongated two-domain molecules with a fairly well conserved core structure (42–45, 56). The receptor “footprint” on arrestins is quite extensive, covering the concave sides of both N- and C-domain (15, 16, 46–48, 57). Receptor-binding arrestin elements fall into two categories: those binding receptor-attached phosphates (30, 55, 58, 59) and residues binding other, non-phosphorylated parts of the receptor (15, 16, 46, 47). Because the phosphates are the common theme, only the latter can discriminate between different GPCR subtypes. The demonstration that swapping two elements between arrestin-1 and -2 completely reverses their receptor preference (15) yielded the first structural clues for GPCR specificity of arrestin proteins. The number of key players in receptor discrimination was recently reduced to ∼10 residues (16). Moreover, it turned out that the elimination of these 10 side chains by alanine substitution in arrestin-1, -2, and -3 essentially destroyed their ability to interact with several GPCRs (16), indicating that these residues are key contributors to the binding energy.
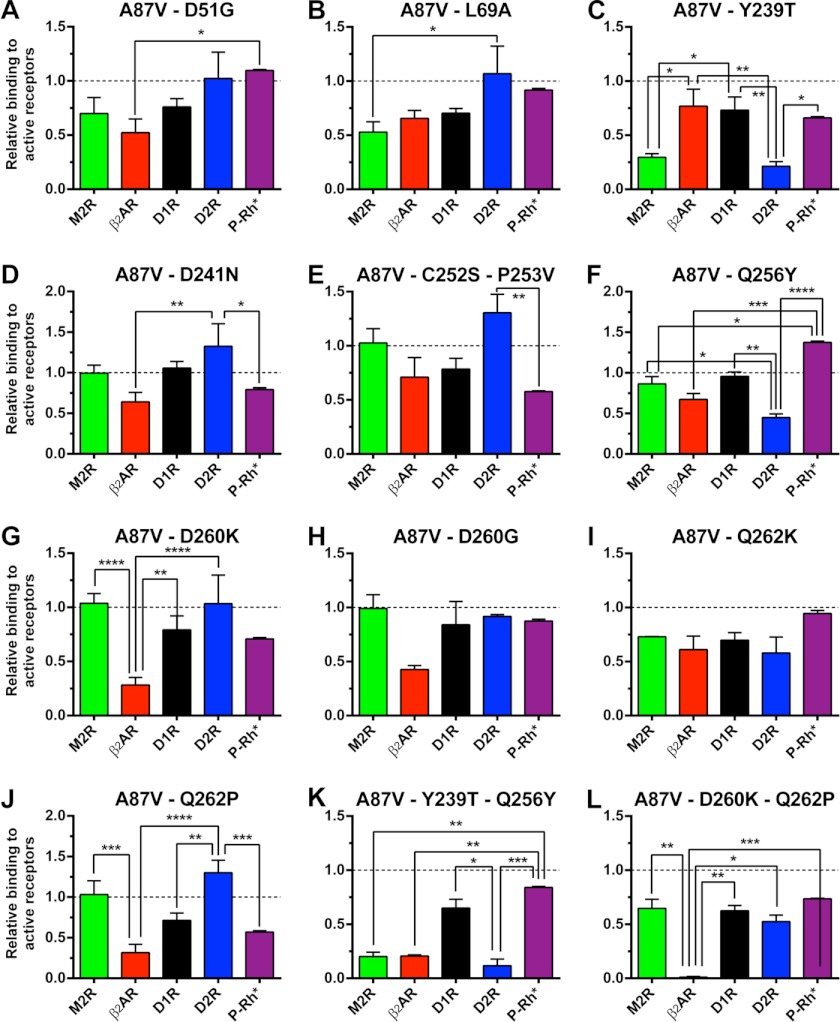
Obviously, placing 20 different amino acids into each of the 10 positions would generate an enormous number of combinations, which is much greater than could be tested experimentally. However, the analysis of arrestin evolution (9) shows that over hundreds of millions of years, very few residues occupied each of these positions (Fig. 1E), which brings the number of meaningful combinations down to manageable. Here we constructed a series of arrestin-3 mutants with specifically targeted receptor discriminator residues. The data show that single substitutions change receptor preference of the inherently promiscuous arrestin-3 up to 4-fold (Fig. 3), whereas double mutations increase the selectivity up to 50–60-fold (Fig. 4). In fact, D260K/Q262P is the first form of any non-visual arrestin that has a dramatic preference for certain receptors over others.
To gauge mutation-specific effects on receptor preference, we compared the relative ability of each form of arrestin-3 to interact with all five receptors, normalized by the binding of A87V base mutant, which was set at 1.0 (Fig. 7). Only one mutation, Q262K, did not significantly affect arrestin selectivity within this group of GPCRs. Mutations L69A and Y239T created bias against M2R; D260G, D260K, and Q262P biased arrestin-3 against β2AR; L69A, D241N, C252S/P253V, and Q262P made arrestin-3 prefer D2R, whereas Y239T and Q256Y biased it against this receptor; finally, Q256Y increased, whereas Q262P decreased, arrestin-3 binding to P-Rh*. Interestingly, four single (Y239T, Q256Y, D260K, and Q262P) and both double mutations (Y239T/Q256Y and D260K/Q262P) yielded complex changes in receptor preference affecting virtually every GPCR tested, each mutation showing its unique pattern of effects (Fig. 7).
FIGURE 7.
Different substitutions of receptor discriminator residues produce unique patterns of changes in arrestin-3 preference for particular GPCRs. To reveal the effects of the indicated mutations, agonist-induced increase in arrestin binding to M2R, β2AR, D1R, and D2R (net BRETMAX in Figs. 2 and 4) and the binding to P-Rh* (Fig. 6) were expressed in relation to those of the base mutant A87V, for which this parameter was set at 1.0 for all receptors. The results for each mutant were analyzed using two-way ANOVA with receptor and arrestin mutation as the main factors, followed by the Holm-Sidak multiple comparison test. The lines above indicate the pairs of values compared, and the stars above the lines indicate statistical significance of the difference, which is indicated as follows. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. Error bars, S.E.
We would like to point out that the choice of these 10 mutations out of many possible (Fig. 1E) was largely based on “educated guessing.” Therefore, it is remarkable that nine of 10 single mutants demonstrated significant changes in receptor preference (Figs. 2, 3, and 7). For example, Y239T shows reduced binding to M2R and D2R but not to the other two assayed receptors. Q256Y binds D2R less than other receptors, and D260K only affects the interaction with β2AR. This 90% success rate suggests that the residues mutated in this study largely determine which GPCRs arrestin binds to. The fact that we were able to achieve 50–60-fold selectivity by combining only two mutations (Fig. 4) further supports our choice of very few “discriminator” residues on an extensive receptor-binding surface of arrestins. Importantly, these proof-of-principle experiments demonstrate the feasibility of constructing non-visual arrestins selective for small groups of GPCRs or even for individual subtypes.
Arrestins are multifunctional proteins. In addition to hundreds of GPCRs, they interact with dozens of other signaling proteins (60), acting as versatile signaling organizers in the cell (9, 61). The binding sites of different partners are structurally separated; receptors engage the concave sides of both domains (15, 16, 46–48, 57), other signaling proteins capable of interacting with the arrestin-receptor complex use the surfaces that do not overlap with receptor-binding elements (62), and trafficking proteins clathrin (63) and AP2 (64) interact with the arrestin C-tail that is detached upon receptor binding (46, 65, 66). This spatial separation creates a possibility to manipulate receptor binding, signaling, and trafficking functions of arrestins independently. Elucidation of the elements responsible for arrestin interactions with individual non-receptor partners enabled the construction of several signaling-biased arrestin mutants. Arrestins with inactivated clathrin and AP2 binding sites suppress GPCR internalization via coated pits (67). The expression of the separated arrestin-2 C-tail, which competes with the arrestin-receptor complex, has the same effect (68). Mutants with the deletion in the interdomain hinge have reduced ability to bind receptors (69, 70) but demonstrate increased affinity for microtubules (70). These mutants recruit ERK2 and ubiquitin ligase Mdm2 to the cytoskeleton, suppressing ERK1/2 phosphorylation in the cell and redirecting the activity of Mdm2 toward microtubule-associated substrates (70). Arrestin-2-R307A binds ERK2 and MEK1 normally but is defective in c-Raf1 binding and therefore fails to facilitate ERK1/2 activation (71). Several arrestin-3 mutants bind all three kinases in the ASK1-MKK4-JNK3 cascade but do not promote JNK3 activation (29), whereas another with enhanced JNK3 binding even acts as a dominant negative, suppressing JNK3 activation by productive scaffolds in the cell (72). Targeted activation of the phosphate sensor of all arrestin subtypes by several mutations yielded enhanced mutants that bind active GPCRs regardless of receptor phosphorylation (24, 25, 55, 73). These mutants suppress the signaling of unphosphorylated receptors in vitro (19), in intact cells (20–22), and even in vivo (23) and dramatically change receptor trafficking (24). The experiments described here are the first demonstration that receptor specificity of inherently promiscuous non-visual arrestins can be significantly narrowed. The development of non-visual arrestins specifically targeting particular GPCRs paves the way to the construction of arrestin mutants linking desired GPCR to signaling pathways of our choosing. Engineered receptor-specific signaling-biased arrestins have obvious potential in research and gene therapy of disorders associated with faulty GPCR signaling.
Supplementary Material
Acknowledgments
We are grateful to Dr. J. A. Javitch (Columbia University, New York) for advice on receptor-arrestin BRET and the plasmid encoding Venus and Dr. N. A. Lambert (Medical College of Georgia, Augusta, GA) for the plasmid encoding Renilla luciferase variant 8.
This work was supported, in whole or in part, by National Institutes of Health Grants GM077561, GM081756, and EY011500 (to V. V. G.).

This article contains supplemental Figs. S1–S7.
We use systematic names of arrestin proteins: arrestin-1 (historic names S-antigen, 48-kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin; for unclear reasons, its gene is called “arrestin 3” in the HUGO database).
- GPCR
- G protein-coupled receptor
- M2R
- M2 muscarinic receptor
- β2AR
- β2-adrenergic receptor
- D1R and D2R
- D1 and D2 dopamine receptor, respectively
- BRET
- bioluminescence resonance energy transfer
- ANOVA
- analysis of variance
- P-Rh*
- phosphorylated light-activated rhodopsin
- arrestin-3-NCA
- arrestin-3-L49A/D51A/R52A/L69A/Y239A/D241A/C252A/P253A/D260A/Q262.
REFERENCES
- 1. Kristiansen K. (2004) Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors. Molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol. Ther. 103, 21–80 [DOI] [PubMed] [Google Scholar]
- 2. Gurevich V. V., Gurevich E. V. (2008) How and why do GPCRs dimerize? Trends Pharmacol. Sci. 29, 234–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gurevich E. V., Tesmer J. J., Mushegian A., Gurevich V. V. (2012) G protein-coupled receptor kinases. More than just kinases and not only for GPCRs. Pharmacol. Ther. 133, 40–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gurevich V. V., Gurevich E. V. (2004) The molecular acrobatics of arrestin activation. Trends Pharmacol. Sci. 25, 105–111 [DOI] [PubMed] [Google Scholar]
- 5. Krupnick J. G., Gurevich V. V., Benovic J. L. (1997) Mechanism of quenching of phototransduction. Binding competition between arrestin and transducin for phosphorhodopsin. J. Biol. Chem. 272, 18125–18131 [DOI] [PubMed] [Google Scholar]
- 6. Wilden U., Hall S. W., Kühn H. (1986) Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc. Natl. Acad. Sci. U.S.A. 83, 1174–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang C. C., Tesmer J. J. (2011) Recognition in the face of diversity. Interactions of heterotrimeric G proteins and G protein-coupled receptor (GPCR) kinases with activated GPCRs. J. Biol. Chem. 286, 7715–7721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mushegian A., Gurevich V. V., Gurevich E. V. (2012) The origin and evolution of G protein-coupled receptor kinases. PLoS One 7, e33806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gurevich E. V., Gurevich V. V. (2006) Arrestins. Ubiquitous regulators of cellular signaling pathways. Genome Biol. 7, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gurevich V. V., Gurevich E. V. (2006) The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol. Ther. 110, 465–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song X., Vishnivetskiy S. A., Seo J., Chen J., Gurevich E. V., Gurevich V. V. (2011) Arrestin-1 expression level in rods. Balancing functional performance and photoreceptor health. Neuroscience 174, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gurevich E. V., Benovic J. L., Gurevich V. V. (2002) Arrestin2 and arrestin3 are differentially expressed in the rat brain during postnatal development. Neuroscience 109, 421–436 [DOI] [PubMed] [Google Scholar]
- 13. Gurevich E. V., Benovic J. L., Gurevich V. V. (2004) Arrestin2 expression selectively increases during neural differentiation. J. Neurochem. 91, 1404–1416 [DOI] [PubMed] [Google Scholar]
- 14. Gurevich V. V., Dion S. B., Onorato J. J., Ptasienski J., Kim C. M., Sterne-Marr R., Hosey M. M., Benovic J. L. (1995) Arrestin interactions with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, β2-adrenergic, and m2 muscarinic cholinergic receptors. J. Biol. Chem. 270, 720–731 [DOI] [PubMed] [Google Scholar]
- 15. Vishnivetskiy S. A., Hosey M. M., Benovic J. L., Gurevich V. V. (2004) Mapping the arrestin-receptor interface. Structural elements responsible for receptor specificity of arrestin proteins. J. Biol. Chem. 279, 1262–1268 [DOI] [PubMed] [Google Scholar]
- 16. Vishnivetskiy S. A., Gimenez L. E., Francis D. J., Hanson S. M., Hubbell W. L., Klug C. S., Gurevich V. V. (2011) Few residues within an extensive binding interface drive receptor interaction and determine the specificity of arrestin proteins. J. Biol. Chem. 286, 24288–24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barak L. S., Ferguson S. S., Zhang J., Caron M. G. (1997) A β-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. J. Biol. Chem. 272, 27497–27500 [DOI] [PubMed] [Google Scholar]
- 18. Schöneberg T., Schulz A., Biebermann H., Hermsdorf T., Römpler H., Sangkuhl K. (2004) Mutant G-protein-coupled receptors as a cause of human diseases. Pharmacol. Ther. 104, 173–206 [DOI] [PubMed] [Google Scholar]
- 19. Gray-Keller M. P., Detwiler P. B., Benovic J. L., Gurevich V. V. (1997) Arrestin with a single amino acid substitution quenches light-activated rhodopsin in a phosphorylation-independent fashion. Biochemistry 36, 7058–7063 [DOI] [PubMed] [Google Scholar]
- 20. Kovoor A., Celver J., Abdryashitov R. I., Chavkin C., Gurevich V. V. (1999) Targeted construction of phosphorylation-independent β-arrestin mutants with constitutive activity in cells. J. Biol. Chem. 274, 6831–6834 [DOI] [PubMed] [Google Scholar]
- 21. Celver J. P., Lowe J., Kovoor A., Gurevich V. V., Chavkin C. (2001) Threonine 180 is required for G-protein-coupled receptor kinase 3- and β-arrestin 2-mediated desensitization of the μ-opioid receptor in Xenopus oocytes. J. Biol. Chem. 276, 4894–4900 [DOI] [PubMed] [Google Scholar]
- 22. Celver J., Vishnivetskiy S. A., Chavkin C., Gurevich V. V. (2002) Conservation of the phosphate-sensitive elements in the arrestin family of proteins. J. Biol. Chem. 277, 9043–9048 [DOI] [PubMed] [Google Scholar]
- 23. Song X., Vishnivetskiy S. A., Gross O. P., Emelianoff K., Mendez A., Chen J., Gurevich E. V., Burns M. E., Gurevich V. V. (2009) Enhanced arrestin facilitates recovery and protects rods lacking rhodopsin phosphorylation. Curr. Biol. 19, 700–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pan L., Gurevich E. V., Gurevich V. V. (2003) The nature of the arrestin x receptor complex determines the ultimate fate of the internalized receptor. J. Biol. Chem. 278, 11623–11632 [DOI] [PubMed] [Google Scholar]
- 25. Gurevich V. V., Pals-Rylaarsdam R., Benovic J. L., Hosey M. M., Onorato J. J. (1997) Agonist-receptor-arrestin, an alternative ternary complex with high agonist affinity. J. Biol. Chem. 272, 28849–28852 [DOI] [PubMed] [Google Scholar]
- 26. Gurevich V. V. (1996) Use of bacteriophage RNA polymerase in RNA synthesis. Methods Enzymol. 275, 382–397 [DOI] [PubMed] [Google Scholar]
- 27. Attramadal H., Arriza J. L., Aoki C., Dawson T. M., Codina J., Kwatra M. M., Snyder S. H., Caron M. G., Lefkowitz R. J. (1992) β-Arrestin2, a novel member of the arrestin/β-arrestin gene family. J. Biol. Chem. 267, 17882–17890 [PubMed] [Google Scholar]
- 28. Sterne-Marr R., Gurevich V. V., Goldsmith P., Bodine R. C., Sanders C., Donoso L. A., Benovic J. L. (1993) Polypeptide variants of β-arrestin and arrestin3. J. Biol. Chem. 268, 15640–15648 [PubMed] [Google Scholar]
- 29. Seo J., Tsakem E. L., Breitman M., Gurevich V. V. (2011) Identification of arrestin-3-specific residues necessary for JNK3 kinase activation. J. Biol. Chem. 286, 27894–27901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gimenez L. E., Kook S., Vishnivetskiy S. A., Ahmed M. R., Gurevich E. V., Gurevich V. V. (2012) Role of receptor-attached phosphates in binding of visual and non-visual arrestins to G protein-coupled receptors. J. Biol. Chem. 287, 9028–9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walther C., Nagel S., Gimenez L. E., Mörl K., Gurevich V. V., Beck-Sickinger A. G. (2010) Ligand-induced internalization and recycling of the human neuropeptide Y2 receptor is regulated by its carboxyl-terminal tail. J. Biol. Chem. 285, 41578–41590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Namkung Y., Dipace C., Javitch J. A., Sibley D. R. (2009) G protein-coupled receptor kinase-mediated phosphorylation regulates post-endocytic trafficking of the D2 dopamine receptor. J. Biol. Chem. 284, 15038–15051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Namkung Y., Dipace C., Urizar E., Javitch J. A., Sibley D. R. (2009) G protein-coupled receptor kinase-2 constitutively regulates D2 dopamine receptor expression and signaling independently of receptor phosphorylation. J. Biol. Chem. 284, 34103–34115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Binkowski B. F., Butler B. L., Stecha P. F., Eggers C. T., Otto P., Zimmerman K., Vidugiris G., Wood M. G., Encell L. P., Fan F., Wood K. V. (2011) A luminescent biosensor with increased dynamic range for intracellular cAMP. ACS Chem. Biol. 6, 1193–1197 [DOI] [PubMed] [Google Scholar]
- 35. Pantel J., Williams S. Y., Mi D., Sebag J., Corbin J. D., Weaver C. D., Cone R. D. (2011) Development of a high throughput screen for allosteric modulators of melanocortin-4 receptor signaling using a real-time cAMP assay. Eur. J. Pharmacol. 660, 139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fan F., Binkowski B. F., Butler B. L., Stecha P. F., Lewis M. K., Wood K. V. (2008) Novel genetically encoded biosensors using firefly luciferase. ACS Chem. Biol. 3, 346–351 [DOI] [PubMed] [Google Scholar]
- 37. Kallal L., Gagnon A. W., Penn R. B., Benovic J. L. (1998) Visualization of agonist-induced sequestration and down-regulation of a green fluorescent protein-tagged β2-adrenergic receptor. J. Biol. Chem. 273, 322–328 [DOI] [PubMed] [Google Scholar]
- 38. Macey T. A., Gurevich V. V., Neve K. A. (2004) Preferential Interaction between the dopamine D2 receptor and Arrestin2 in neostriatal neurons. Mol. Pharmacol. 66, 1635–1642 [DOI] [PubMed] [Google Scholar]
- 39. Macey T. A., Liu Y., Gurevich V. V., Neve K. A. (2005) Dopamine D1 receptor interaction with arrestin3 in neostriatal neurons. J. Neurochem. 93, 128–134 [DOI] [PubMed] [Google Scholar]
- 40. Kohout T. A., Lin F. S., Perry S. J., Conner D. A., Lefkowitz R. J. (2001) β-Arrestin 1 and 2 differentially regulate heptahelical receptor signaling and trafficking. Proc. Natl. Acad. Sci. U.S.A. 98, 1601–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oakley R. H., Laporte S. A., Holt J. A., Caron M. G., Barak L. S. (2000) Differential affinities of visual arrestin, β-arrestin1, and β-arrestin2 for G protein-coupled receptors delineate two major classes of receptors. J. Biol. Chem. 275, 17201–17210 [DOI] [PubMed] [Google Scholar]
- 42. Hirsch J. A., Schubert C., Gurevich V. V., Sigler P. B. (1999) The 2.8 Å crystal structure of visual arrestin. A model for arrestin's regulation. Cell 97, 257–269 [DOI] [PubMed] [Google Scholar]
- 43. Han M., Gurevich V. V., Vishnivetskiy S. A., Sigler P. B., Schubert C. (2001) Crystal structure of β-arrestin at 1.9 Å. Possible mechanism of receptor binding and membrane translocation. Structure 9, 869–880 [DOI] [PubMed] [Google Scholar]
- 44. Milano S. K., Pace H. C., Kim Y. M., Brenner C., Benovic J. L. (2002) Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry 41, 3321–3328 [DOI] [PubMed] [Google Scholar]
- 45. Zhan X., Gimenez L. E., Gurevich V. V., Spiller B. W. (2011) Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J. Mol. Biol. 406, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hanson S. M., Francis D. J., Vishnivetskiy S. A., Kolobova E. A., Hubbell W. L., Klug C. S., Gurevich V. V. (2006) Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 103, 4900–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hanson S. M., Gurevich V. V. (2006) The differential engagement of arrestin surface charges by the various functional forms of the receptor. J. Biol. Chem. 281, 3458–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ohguro H., Palczewski K., Walsh K. A., Johnson R. S. (1994) Topographic study of arrestin using differential chemical modifications and hydrogen/deuterium exchange. Protein Sci. 3, 2428–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jagadeesh G., Cragoe E. J., Jr., Deth R. C. (1990) Modulation of bovine aortic α-2 receptors by Na+, 5′-guanylylimidodiphosphate, amiloride, and ethylisopropylamiloride. Evidence for receptor G-protein precoupling. J. Pharmacol. Exp. Ther. 252, 1184–1196 [PubMed] [Google Scholar]
- 50. Frances B., Puget A., Moisand C., Meunier J. C. (1990) Apparent precoupling of κ- but not μ-opioid receptors with a G protein in the absence of agonist. Eur. J. Pharmacol. 189, 1–9 [DOI] [PubMed] [Google Scholar]
- 51. Tian W. N., Duzic E., Lanier S. M., Deth R. C. (1994) Determinants of α2-adrenergic receptor activation of G proteins. Evidence for a precoupled receptor/G protein state. Mol. Pharmacol. 45, 524–531 [PubMed] [Google Scholar]
- 52. Galés C., Rebois R. V., Hogue M., Trieu P., Breit A., Hébert T. E., Bouvier M. (2005) Real-time monitoring of receptor and G-protein interactions in living cells. Nat. Methods 2, 177–184 [DOI] [PubMed] [Google Scholar]
- 53. Hein P., Frank M., Hoffmann C., Lohse M. J., Bünemann M. (2005) Dynamics of receptor/G protein coupling in living cells. EMBO J. 24, 4106–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gurevich V. V., Benovic J. L. (1993) Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J. Biol. Chem. 268, 11628–11638 [PubMed] [Google Scholar]
- 55. Gurevich V. V., Benovic J. L. (1995) Visual arrestin binding to rhodopsin. Diverse functional roles of positively charged residues within the phosphorylation recognition region of arrestin. J. Biol. Chem. 270, 6010–6016 [DOI] [PubMed] [Google Scholar]
- 56. Sutton R. B., Vishnivetskiy S. A., Robert J., Hanson S. M., Raman D., Knox B. E., Kono M., Navarro J., Gurevich V. V. (2005) Crystal structure of cone arrestin at 2.3 Å. Evolution of receptor specificity. J. Mol. Biol. 354, 1069–1080 [DOI] [PubMed] [Google Scholar]
- 57. Pulvermüller A., Schroder K., Fischer T., Hofmann K. P. (2000) Interactions of metarhodopsin II. Arrestin peptides compete with arrestin and transducin. J. Biol. Chem. 275, 37679–37685 [DOI] [PubMed] [Google Scholar]
- 58. Gurevich V. V., Benovic J. L. (1997) Mechanism of phosphorylation recognition by visual arrestin and the transition of arrestin into a high affinity binding state. Mol. Pharmacol. 51, 161–169 [DOI] [PubMed] [Google Scholar]
- 59. Vishnivetskiy S. A., Paz C. L., Schubert C., Hirsch J. A., Sigler P. B., Gurevich V. V. (1999) How does arrestin respond to the phosphorylated state of rhodopsin? J. Biol. Chem. 274, 11451–11454 [DOI] [PubMed] [Google Scholar]
- 60. Xiao K., McClatchy D. B., Shukla A. K., Zhao Y., Chen M., Shenoy S. K., Yates J. R., 3rd, Lefkowitz R. J. (2007) Functional specialization of β-arrestin interactions revealed by proteomic analysis. Proc. Natl. Acad. Sci. U.S.A. 104, 12011–12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. DeWire S. M., Ahn S., Lefkowitz R. J., Shenoy S. K. (2007) β-Arrestins and cell signaling. Annu. Rev. Physiol. 69, 483–510 [DOI] [PubMed] [Google Scholar]
- 62. Gurevich V. V., Gurevich E. V. (2010) Custom-designed proteins as novel therapeutic tools? The case of arrestins. Expert Rev. Mol. Med. 12, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Goodman O. B., Jr., Krupnick J. G., Santini F., Gurevich V. V., Penn R. B., Gagnon A. W., Keen J. H., Benovic J. L. (1996) β-Arrestin acts as a clathrin adaptor in endocytosis of the β2-adrenergic receptor. Nature 383, 447–450 [DOI] [PubMed] [Google Scholar]
- 64. Laporte S. A., Oakley R. H., Zhang J., Holt J. A., Ferguson S. S., Caron M. G., Barak L. S. (1999) The β2-adrenergic receptor/β-arrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc. Natl. Acad. Sci. U.S.A. 96, 3712–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vishnivetskiy S. A., Francis D., Van Eps N., Kim M., Hanson S. M., Klug C. S., Hubbell W. L., Gurevich V. V. (2010) The role of arrestin α-helix I in receptor binding. J. Mol. Biol. 395, 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhuang T., Vishnivetskiy S. A., Gurevich V. V., Sanders C. R. (2010) Elucidation of inositol hexaphosphate and heparin interaction sites and conformational changes in arrestin-1 by solution nuclear magnetic resonance. Biochemistry 49, 10473–10485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim Y. M., Benovic J. L. (2002) Differential roles of arrestin-2 interaction with clathrin and adaptor protein 2 in G protein-coupled receptor trafficking. J. Biol. Chem. 277, 30760–30768 [DOI] [PubMed] [Google Scholar]
- 68. Orsini M. J., Benovic J. L. (1998) Characterization of dominant negative arrestins that inhibit β2-adrenergic receptor internalization by distinct mechanisms. J. Biol. Chem. 273, 34616–34622 [DOI] [PubMed] [Google Scholar]
- 69. Vishnivetskiy S. A., Hirsch J. A., Velez M. G., Gurevich Y. V., Gurevich V. V. (2002) Transition of arrestin into the active receptor-binding state requires an extended interdomain hinge. J. Biol. Chem. 277, 43961–43967 [DOI] [PubMed] [Google Scholar]
- 70. Hanson S. M., Cleghorn W. M., Francis D. J., Vishnivetskiy S. A., Raman D., Song X., Nair K. S., Slepak V. Z., Klug C. S., Gurevich V. V. (2007) Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J. Mol. Biol. 368, 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Coffa S., Breitman M., Spiller B. W., Gurevich V. V. (2011) A single mutation in arrestin-2 prevents ERK1/2 activation by reducing c-Raf1 binding. Biochemistry 50, 6951–6958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Breitman M., Kook S., Gimenez L. E., Lizama B. N., Palazzo M. C., Gurevich E. V., Gurevich V. V. (2012) Silent scaffolds. Inhibition of c-Jun N-terminal kinase 3 activity in cell by dominant-negative arrestin-3 mutant. J. Biol. Chem. 287, 19653–19664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gurevich V. V. (1998) The selectivity of visual arrestin for light-activated phosphorhodopsin is controlled by multiple nonredundant mechanisms. J. Biol. Chem. 273, 15501–15506 [DOI] [PubMed] [Google Scholar]
- 74. Gurevich V. V., Benovic J. L. (1992) Cell-free expression of visual arrestin. Truncation mutagenesis identifies multiple domains involved in rhodopsin interaction. J. Biol. Chem. 267, 21919–21923 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.