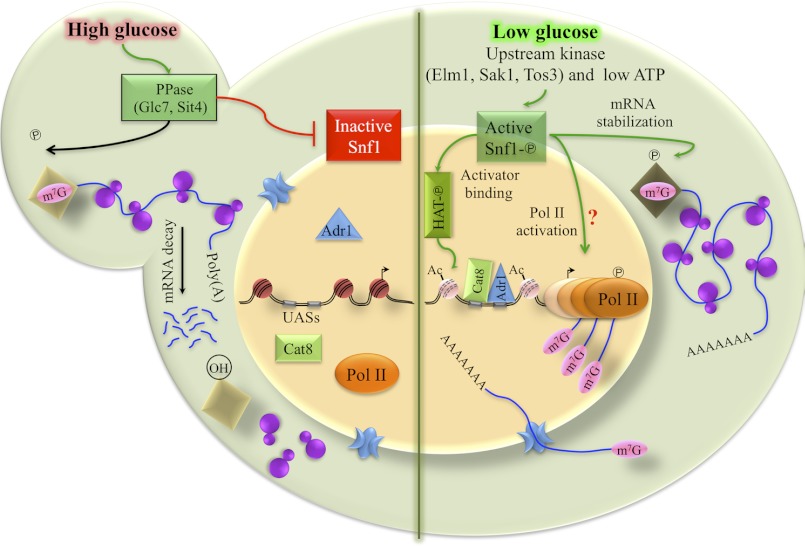
FIGURE 7.
Model of Snf1 regulation of Adr1-dependent gene expression. Activation of and by Snf1 is indicated by green arrows; red arrows indicate pathways or functions that inactivate Snf1 and Snf1-dependent functions. In brief, activation of Snf1 occurs through phosphorylation of Snf1 on Thr-210, a process that is reversed in the presence of glucose through the action of two protein phosphatases, Glc7 and Sit4. The model depicts active Snf1 stimulating acetylation of nucleosomal histones, a process that could occur by directly stimulating a histone acetyltransferase (as shown), by inactivating a histone deacetylase, or by phosphorylation-dependent acetylation of histones. Once bound, Adr1 and Cat8 stimulate promoter nucleosome remodeling and PIC recruitment. A subsequent step in PIC activation is Snf1-dependent as is stabilization of cytoplasmic mRNA stability. The latter process could occur through protection of the 5′ end of glucose-repressed mRNAs, by altering cytoplasmic mRNA decay processes, or through some other unknown mechanism. The protection is represented by a phosphorylated diamond protecting the 5′ cap of a mRNA. Snf1 may also affect transcription elongation because RNA pol II is engaged in the ORF, but transcript accumulation is not observed after Snf1 is inhibited. The green arrow showing Snf1 affecting this process has a question mark (?) because we have not directly demonstrated an effect on transcript elongation.