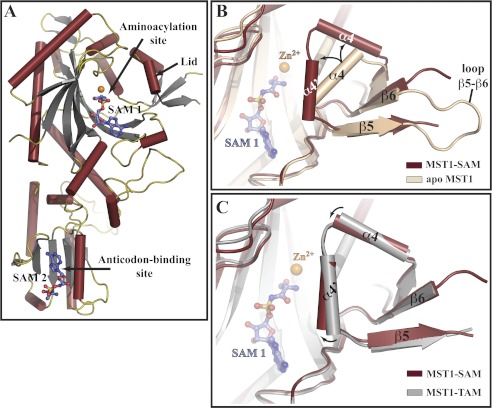
FIGURE 4.
Ser-AMP analog binds to two sites in MST1 and stabilizes the closed conformation of the aminoacylation domain. A, ribbon diagram of the crystal structure of the MST1-SAM binary complex determined at 2.87 Å resolution. SAM 1 is bound to the aminoacylation site along with a Zn2+ ion (orange sphere), whereas SAM 2 binds to the site in the anticodon-binding domain implicated in the anticodon sequence recognition (see supplemental Figs. S4 and S5 for more details). Helices, strands, and loops are dark red, gray, and olive, respectively. SAM molecules are shown as blue sticks. B, superimpositioning of the apo-MST1 structure (beige; PDB ID code 3UGQ) onto MST1-SAM (dark red) reveals a structural rearrangement of the active-site lid. In particular, loop β5-β6 becomes disordered, and helix α4 breaks into two smaller helices (labeled here as α4 and α4′), which are now positioned at an angle of ∼90°. C, comparison of the crystal structures of MST1-SAM (dark red) and MST1-TAM (gray; PDB ID code 3UH0) reveals that helices α4 and α4′ move closer to the active site when MST1 binds SAM. SAM (blue balls-and-sticks) and Zn2+ (orange sphere) are shown as reference points.