Figure 2. Immunoblot analysis for FimA using whole-cell sonicates.
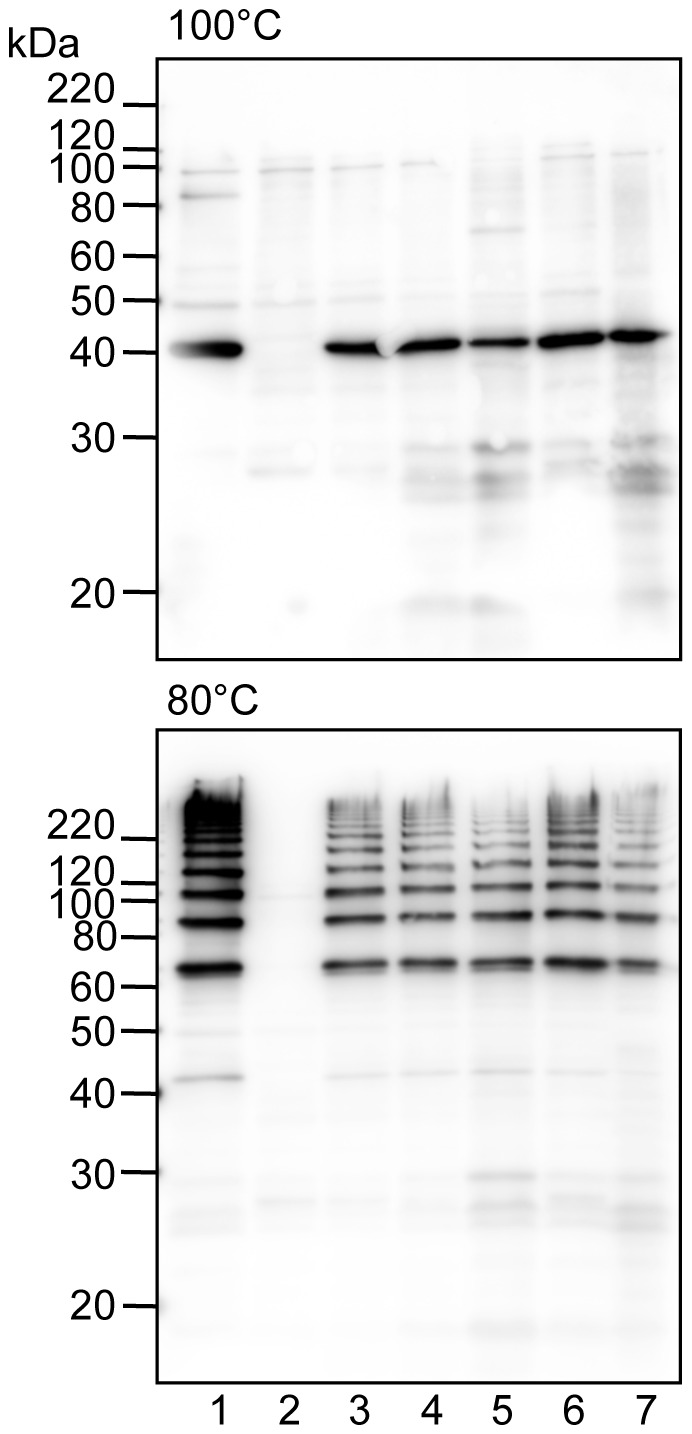
Whole-cell sonicates were denatured in an SDS-containing buffer with 2-mercaptoethanol by heating at 100°C (upper) and 80°C (lower) for 10 min, then subjected to SDS-PAGE and immunoblot analysis. A mixture of specific antisera to the FimA polymer and monomer was used. Antigen samples were as follows: P. gingivalis ATCC 33277 Δmfa1 (expresses native FimA fimbriae, lane 1), P. gingivalis ATCC 33277 Δmfa1 Δfim cluster (FimA deficient, lane 2) with fimX-pgmA-fimA (lane 3), pgmA-fimA (lane 4), fimA (lane 5), fimX & fimA (lane 6) complementarily introduced, and P. gingivalis W83 Δfim cluster with fimA introduced (lane 7). All introduced genes originated from P. gingivalis ATCC 33277. Incomplete dissociation of FimA polymers produces a ladder-like band indicating oligomers in the lower panel. Note that bands slightly higher than 60 kDa (the lower panel) are dimers although monomers appear to be about 40 kDa (the upper panel).
