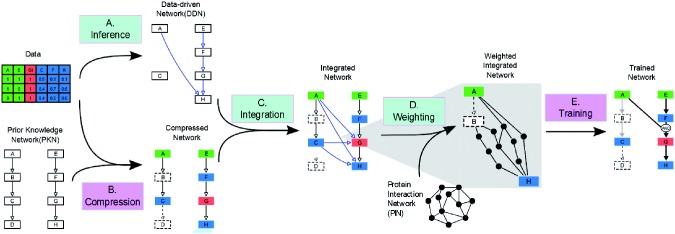
Fig. 1.
Integrated pipeline of CNORfeeder (light boxes) and CellNOptR (dark boxes). (A) Data are used to infer, using reverse-engineering methods, a strictly DDN; (B) the PKN is compressed according to the data (dark, middle and light grey nodes are, respectively, stimulated, inhibited and measured), removing non-identifiable nodes (dashed); (C) the compressed network is integrated with the DDN (dotted links are obtained from the DDN and continuous links from the PKN); (D) information derived from PINs are used to support and prioritize integrated links; (E) The integrated network is used as input for the training: in the trained model, thick black lines denote interactions (and gates) in the trained model, and light-grey links denote presence in the integrated network but not in the trained model