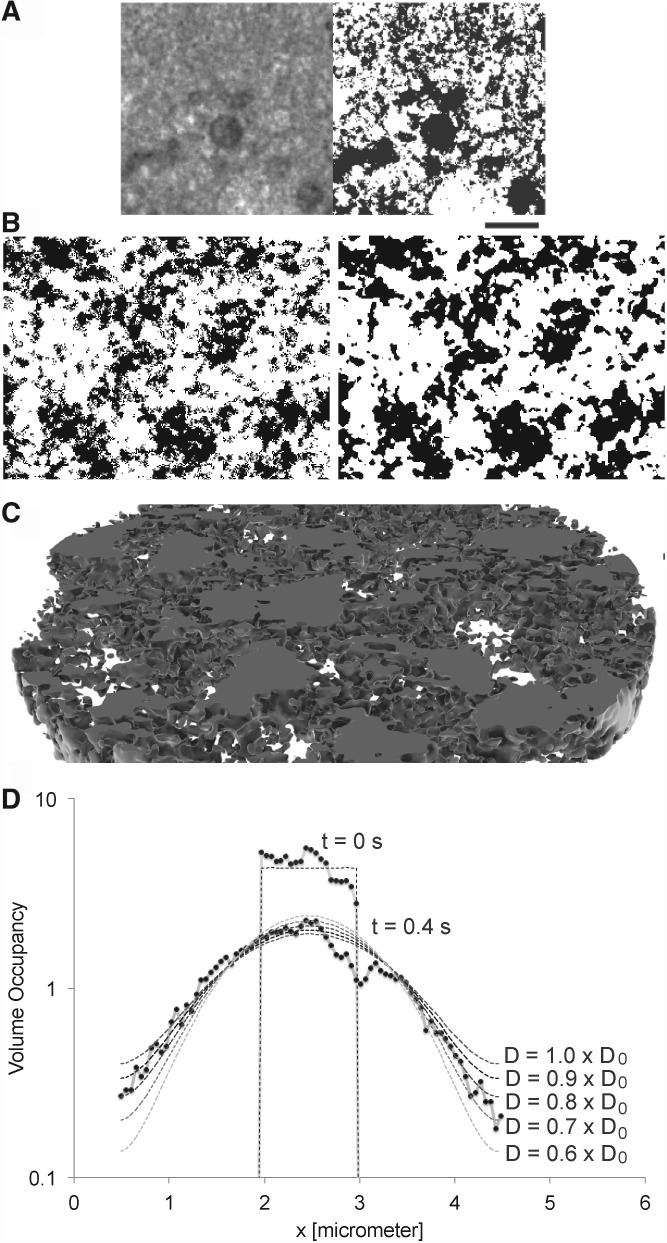
Fig. 6.
A Based on the statistics of binarized Transmission electron microscopy (TEM) images from (Hiroi et al., 2011) B realistically looking intracellular obstacle geometries were generated (Hiroi et al., 2012). C shows a 3D section of a generated volume, which looks similar like other 3D observations of membrane enclosed compartments like e.g. ER or Golgi (rendered with BioInspire from ScienceVisuals). D Diffusion through such a structured volume, simulated on the Gillespie level. Apparently at x = 3μm a high obstacle density leads to a lower particle density