Fig. 2.
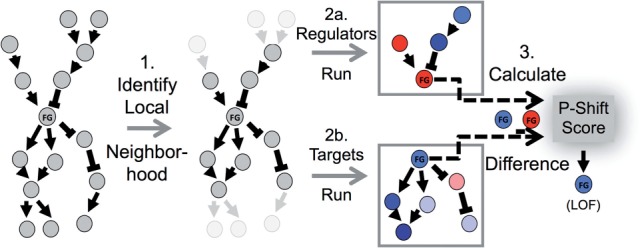
Overview of the PARADIGM-SHIFT method. Inference is centered on a FG for which mutations have been detected in one or more samples. First, a local neighborhood around FG is isolated from the full pathway. PARADIGM is run in two modes using only the local neighborhood. A ‘Regulators Run’ infers the activity of FG using only its upstream regulators in the local neighborhood; a ‘Targets Run’ infers FG's activity using only downstream targets. Finally, the difference between these two runs is calculated and used as the P-Shift Score. In this case, LOF is predicted because the downstream targets indicate lower activity than the upstream regulators of FG
