Fig. 1.
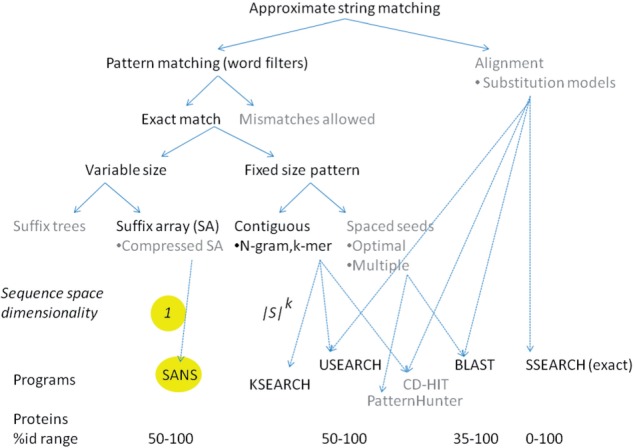
A classification of approaches used by representative application programs for protein sequence retrieval. This work focuses on word filters based on suffix arrays or k-mer counting. Related approaches or data structures are shown in gray and not expanded. SANS is simple and fast but has a limited application range. Other fast programs calculate explicit alignments for a filtered subset of database proteins. SSEARCH calculates the optimal alignment between the query and all database proteins
