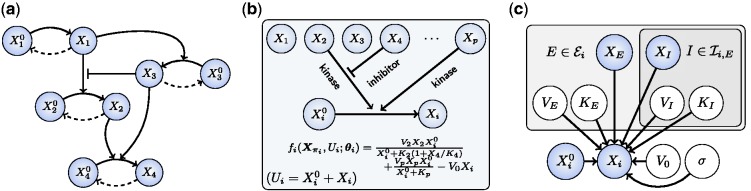
Fig. 1.
Overview of approach. (a) An example of a phosphorylation network. (b) Our approach couples automatic generation of chemical models with Bayesian model selection to infer regulators πi of species i. (c) A statistical formulation (graphical model) for equilibrium phosphorylation of species i is characterized by specifying kinases (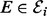 ) and inhibitors (
) and inhibitors ( ) of kinases. [Bounding boxes are used to indicate multiplicity of variables, shaded nodes are observed with noise.]
) of kinases. [Bounding boxes are used to indicate multiplicity of variables, shaded nodes are observed with noise.]