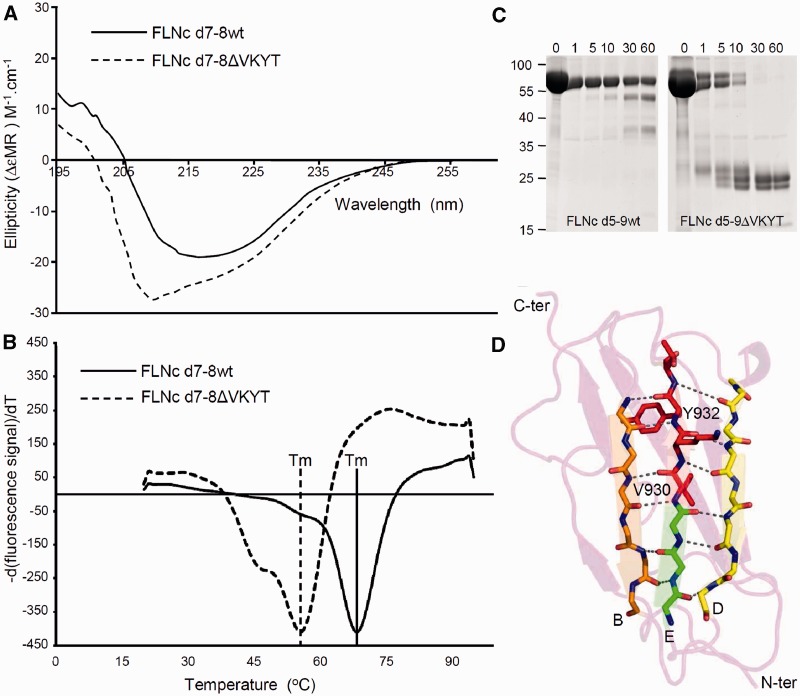
Figure 8.
Biophysical characterization and limited proteolysis of the FLNC p.V930_T933del mutation. (A) Circular dichroism spectroscopy performed with purified, bacterially expressed wild-type and deletion mutant FLNC d7-8ΔVKYT constructs. The mean residue ellipticity (ΔεMR) of the wild-type shows a minimum in the spectrum at 218 nm and a positive value at 205 nm, while the deletion mutant displays a blue shift with minimum at 208 nm and positive ellipticity at 200 nm, a characteristic sign of the presence of unstructured regions. Solid line = wild-type; dashed line = FLNC d7-8ΔVKYT construct. (B) Differential scanning fluorimetry. Temperature denaturation experiments for FLNC d7–8 and deletion mutant FLNC d7–8ΔVKYT. The melting temperature (Tm) is determined at the inflection point of the fluorescence signal, the first derivative of which is reported here. Solid line = wild-type; dashed line = deletion mutant, scaled to the wild-type. (C) Protein stability analysed by thermolysin digestion. Wild-type and mutant FLNC d5-9ΔVKYT were treated with the protease thermolysin for 1 to 60 min, as indicated above each lane. Samples were analysed by PAGE. Note that the mutant protein was completely digested after 30 min, while a significant portion of the wild-type variant was still intact after 60 min of incubation, indicating less stable folding of the mutant protein. (D) Ribbon diagram of homology model of immunoglobulin-like domain 7 of human FLNC. β-strands B, E and D forming a β-sheet are coloured orange, green and yellow, respectively. Main chain atoms of residues involved hydrogen bonds indicated by dashed lines, stabilizing the β-sheet are depicted in stick model, with N and O coloured in blue and red. Residues 930VKYT933 that are missing in the deletion mutant are shown as full-atom stick model and coloured in red.