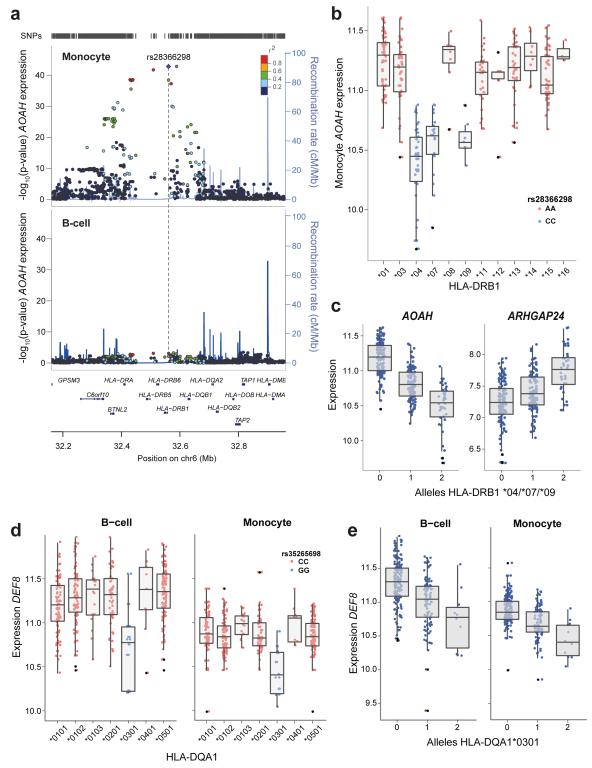
Figure 5. Imputation of HLA status resolves cell-specific trans-associated gene expression to carriage of specific classical HLA alleles.
(a) Regional association plots from monocyte and B-cell data demonstrating the monocyte specific trans association of the chromosome 7 gene AOAH, to the class II MHC gene HLA-DRB1. There is no association in B-cell dataset, whilst the peak eSNP from monocyte dataset is rs28366298 (pmonocyte=1.6×10−43). (b) Imputation of class II alleles to 2 digit resolution demonstrates that the C allele of rs28366298 is specific to DRB1 *04, *07 and *09 alleles and these alleles are associated with reduced AOAH expression. For clarity only homozygotes are plotted with two AOAH expression values per individual (corresponding to each DRB allele) are displayed. (c) The number of DRB1 *04/*07/*09 alleles carried by an individual is significantly associated with reduced expression of AOAH (pB-cell <2.2×10−16, one-way ANOVA) and increased expression of ARHGAP24 (pmonocyte=3.0×10−14, one-way ANOVA). (d) Imputation of DQA1 status to 4 digits demonstrates that the G allele of rs35265698 is significantly associated with reduced expression of DEF8 in both B-cells and monocytes (pB-cell=6.2×10−13, pmonocyte=9.0×10−17). This allele is unique to DQA1*0301 as illustrated. For clarity only homozygotes of rs35265698 are plotted with two DEF8 expression values per individual (corresponding to each DQA allele) displayed. (e) Higher numbers of DQA1*0301 alleles possessed by an individual is significantly associated with reduced DEF8 expression (pB-cell=6.2×10−13, pmonocyte <2.2×10−16)