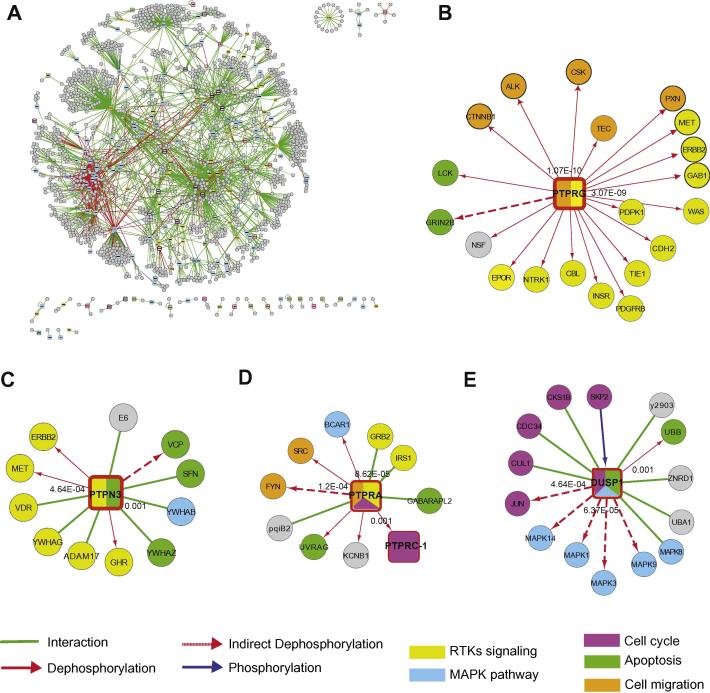
Fig. 4.
The human phosphatase interactome. (A) Graph representation of the human phosphatase interactome. 2496 protein–protein interaction pairs involving one of the phosphatase proteins were retrieved from the HomoMINT and IntAct databases [43,44]. The network was generated with the Cytoscape graphic software. Squared nodes represent phosphatases, while rounded grey nodes represent their targets. The phosphatase color code reflects class affiliation according to the color code convention used throughout this review. Interactors (circles) of “cancer PTPs” whose biological process is not annotated in UniProtKB with GO terms (B) PTPRG-B, (C) PTPN3, (D) PTPRA and (E) DUSP1) were extracted from the global phosphatase interactome and marked in different colors, according to their functional association with GO-Biological Process terms. The phosphatase square nodes are labeled with sectors whose color matches that of the GO-Biological Process term that was significantly overrepresented in the phosphatase interactors and substrates (p-value < 0.002). Edges are colored according to the functional relationships between the nodes they connect: physical associations, dephosphorylations in red and phosphorylation reactions in green and blue respectively. Continuous and dashed lines represent direct (demonstrated in vitro with purified proteins) and indirect dephosphorylation (demonstrated in vivo), respectively.