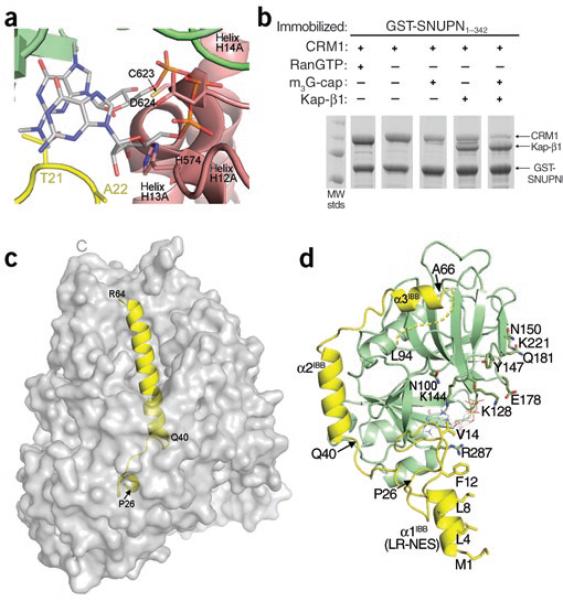
Figure 3.
Comparison of CRM1-, Kap-β1- and m3G-cap-nucleotide-bound SNUPN proteins. (a) The m3G-cap binding site in the CRM1–SNUPN complex. The m3G-cap nucleotide from the superimposed structure of nucleotide-bound NBD is shown as sticks (PDB 1XK5)13. CRM1 is shown as a pink ribbon and the sIBB and NBD of SNUPN as yellow and green ribbons, respectively. (b) Pulldown assays using immobilized GSTSNUPN and CRM1 were performed in the presence of RanGTP, m3G-cap nucleotide or Kap-β1. Bound proteins were separated by SDS-PAGE and visualized with Coomassie blue staining. (c) The Kap-β1–sIBB complex (PDB 2P8Q) is shown with the molecular surface of Kap-β1 in gray and the sIBB as a yellow ribbon. (d) Ribbon diagram of CRM1-bound SNUPN, showing that most of the sIBB (yellow) interacts with its NBD (green). m3G-cap nucleotide from the superimposed structure of nucleotide-bound NBD13 is shown as sticks. The structure is drawn with residues 40–52 of the sIBB in the same orientation as in c.