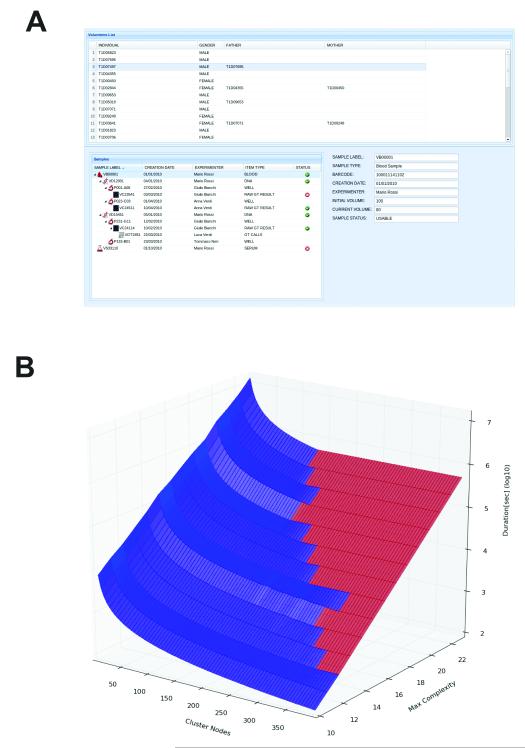
Figure 5. Non-Image Data and the Use of Metadata for Large-Scale Computations.
(A) Alternative use of OMERO for storing and tracking clinical laboratory specimens and genotype data. Sample type, creation date, processing, assay results, genotyping results, and any familial relationships are all recorded. The modelling for this system is based on OpenEHR archetypes. See Supplemental Note for more details.
(B) Performance of a SNP imputing calculation as a function of pedigree complexity (an index of the number of members of a genetically related family) and compute nodes used to perform the calculation. Low complexity imputation benefits from increasing the number of compute nodes, and only occupies nodes for relatively short periods. High complexity calculations take orders of magnitude longer, and do not benefit from adding more nodes. The point where adding number of nodes provides no improvement in performance is shown by the interface between the blue and red domains. Time and cost-efficient use of compute resources therefore depends on access to metadata, determination of complexity, and planning of processing strategy before initiating the imputing calculation.