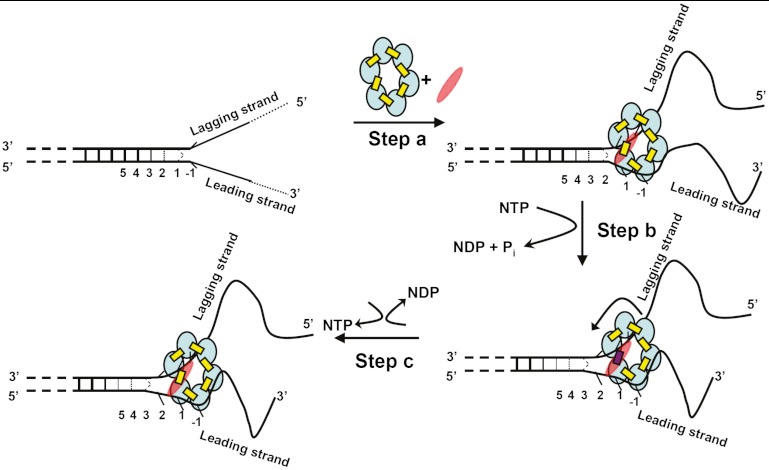
Fig. 5.
Proposed unwinding mechanism of the T4 primosome helicase. The constituents of the primosome are gp41 subunits (blue ellipses), gp61 subunit (red ellipses), GTP (yellow rectangles), and GDP (red rectangles). The “degree of openness” of the base pairs adjacent to the ss–dsDNA junction is indicated in each panel, and the numbers below each DNA construct represent the numbering of the various base pairs prior to initial helicase binding. Step A: The GTP-bound gp41 hexameric helicase loads onto the free DNA fork construct and the gp61 primase subunit binds and stabilizes the complex at the fork junction. Positioning is facilitated by the uniquely unstacked conformation of the -1 bases. As a result of this initial binding the first duplex (breathing) base pair at position 1 is fully unwound and the breathing of the base pairs at initial positions 2, 3, and 4 are all enhanced. Step B: GTP hydrolysis occurs at the gp41–gp41 interface positioned adjacent to the bound gp61 subunit, destabilizing that subunit interface and permitting the primosome to “capture” the now unwound first base pair of the original duplex. This base pair becomes the new -1 position, thereby moving the breathing properties of each base pair at the fork one position further into the duplex sequence. The gp41 hexamer rotates by one subunit (approximately 60°) and the primase translocates to the next gp41–gp41 interface. Step C: The GDP (and Pi) hydrolysis products formed in step B dissociate, a new GTP binds and stabilizes the previously destabilized gp41–gp41 subunit interface, and the primosome helicase is ready to begin a new unwinding-rotation-hydrolysis cycle.