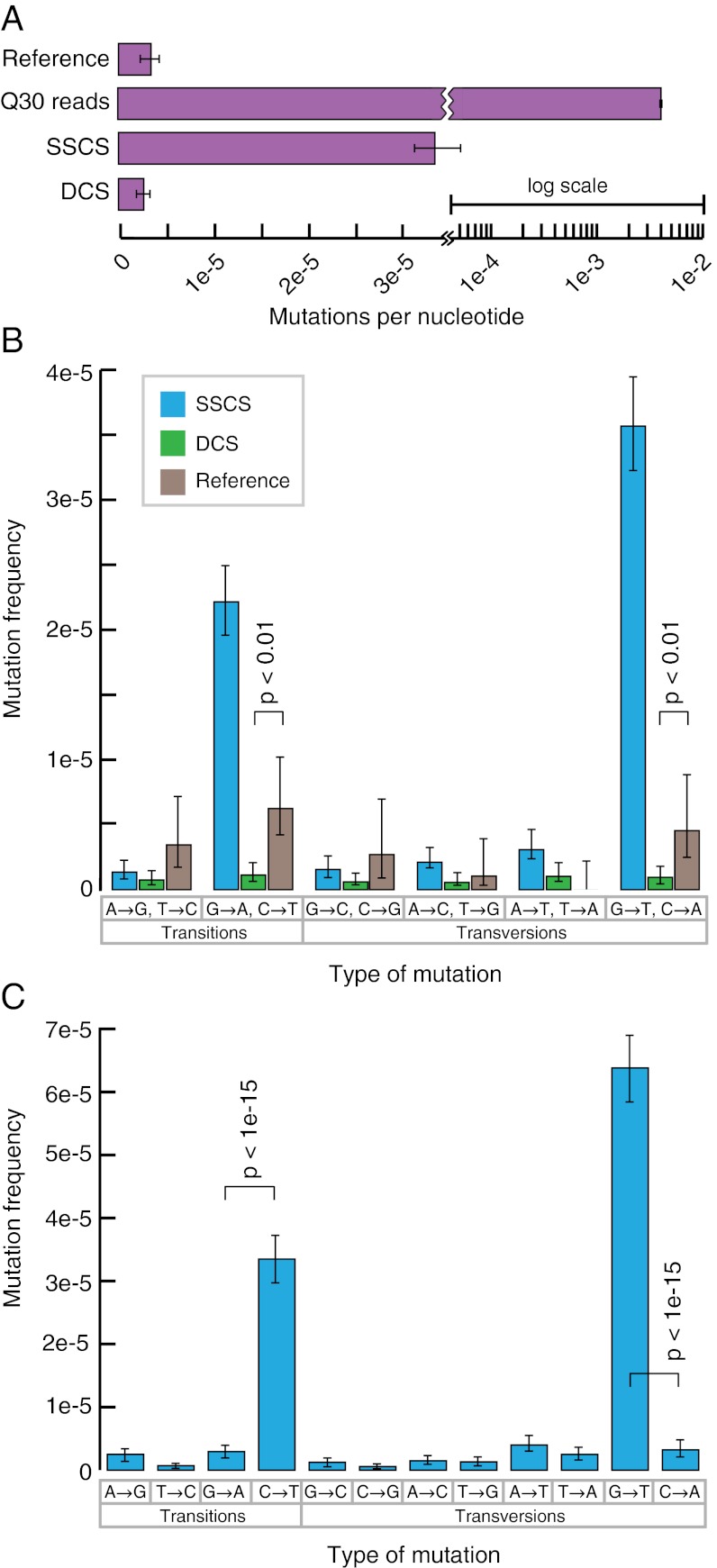
Fig. 2.
Duplex Sequencing of M13mp2 DNA. (A) Average mutation frequency of M13mp2 DNA as measured by a standard sequencing approach, SSCS, and DCS. Reference value of 3.0 × 10−6 is from ref. 34. Note that the axis is plotted on a split-log scale. (B) Single-strand consensus sequences (SSCSs) reveal a large excess of G→A/C→T and G→T/C→A mutations, whereas duplex consensus sequences (DCSs) yield a balanced spectrum. Mutation frequencies are grouped into reciprocal mispairs, as DCS analysis only scores mutations present in both strands of duplex DNA. All significant (P < 0.05) differences between DCS analysis and the literature reference values are noted. (C) Complementary types of mutations should occur at approximately equal frequencies within a DNA fragment population derived from duplex molecules. However, SSCS analysis yields a 15-fold excess of G→T mutations relative to C→A mutations and an 11-fold excess of C→T mutations relative to G→A mutations. All significant (P < 0.05) differences between paired reciprocal mutation frequencies are noted.