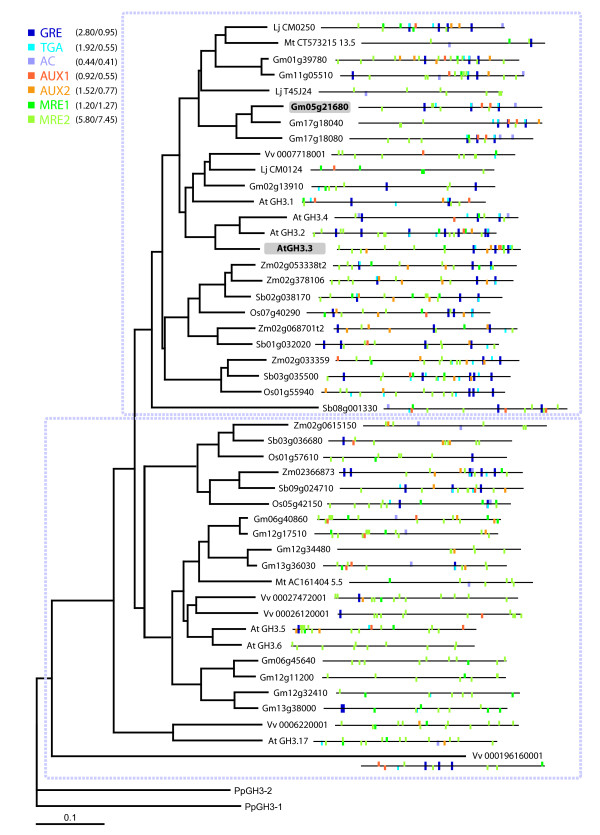
Figure 1.
Phylogram of GH3 homologs from various species and their associated promoter cis-element organisations. The nearest neighbours of the well-characterized soybean GH3 (Gm05g21680) [36] from several plant species were compiled and the corresponding predicted protein sequences were rooted to Physcomitrella patens PpGH3-1 (at the bottom) to create a neighbour-joining phylogram. The −1000 bp promoter sequences of the corresponding GH3 genes were manually co-plotted, in 5’ to 3’ orientation onto the phylogram. The location of specific ZRE, MRE and AuxRE cis-elements (see Table 1) are given using the presented colour code. The indicated boxes divide the phylogram into the ZRE-rich (upper section) and ZRE-poor (lower section) clade. The average number of cis-elements per promoter within the clades (ZRE rich/ZRE poor) is given next to the motif colour code. The position of the soybean GH3 (GmGH3) and its related Arabidopsis homolog (AtGH3.3), used in this study is highlighted in grey. The indicated scale reflects the number of amino acid substitutions per site.