Abstract
The mPER1 and mPER2 proteins have important roles in the circadian clock mechanism, whereas mPER3 is expendable. Here we examine the posttranslational regulation of mPER3 in vivo in mouse liver and compare it to the other mPER proteins to define the salient features required for clock function. Like mPER1 and mPER2, mPER3 is phosphorylated, changes cellular location, and interacts with other clock proteins in a time-dependent manner. Consistent with behavioral data from mPer2/3 and mPer1/3 double-mutant mice, either mPER1 or mPER2 alone can sustain rhythmic posttranslational events. However, mPER3 is unable to sustain molecular rhythmicity in mPer1/2 double-mutant mice. Indeed, mPER3 is always cytoplasmic and is not phosphorylated in the livers of mPer1-deficient mice, suggesting that mPER3 is regulated by mPER1 at a posttranslational level. In vitro studies with chimeric proteins suggest that the inability of mPER3 to support circadian clock function results in part from lack of direct and stable interaction with casein kinase Iɛ (CKIɛ). We thus propose that the CKIɛ-binding domain is critical not only for mPER phosphorylation but also for a functioning circadian clock.
Circadian rhythms have been observed in organisms from cyanobacteria to humans (11). These rhythms are under the direct influence of environmental cues, most notably the day-night cycles and by a genetically determined, endogenous clock (2, 6). In mammals, a master clock located in the suprachiasmatic nuclei of the anterior hypothalamus controls peripheral clocks through neuronal and humoral connections (19).
A major area of interest in circadian biology is to understand the time-keeping mechanism at the molecular level. Transcriptional feedback loops constitute a central feature of most, if not all, circadian clocks (2, 6, 18). In the mouse, two basic-helix-loop-helix-PAS-containing transcription factors, CLOCK and BMAL1, form heterodimers that bind to E-box enhancer sequences and drive the rhythmic transcription of three Period genes (mPer1, mPer2, and mPer3) and two Cryptochrome genes (mCry1 and mCry2) (9, 12). As mPER and mCRY are translated in the cytoplasm, they form mPER-mCRY complexes, which then translocate into the nucleus to inhibit their own transcription by directly interacting with the CLOCK-BMAL1 complex (14, 15, 17, 21). Based on in vitro and in vivo studies, it appears that the CRY proteins play a major role in this inhibition (8, 10, 14, 21).
At the posttranslational level, the mPER1 and mPER2 proteins are temporally phosphorylated in a circadian manner (15). Casein kinase Iɛ (CKIɛ) and CKIδ are important kinases for PER and possibly other clock proteins in mammals (4, 7, 15, 16). Phosphorylation of clock proteins may affect their stability, cellular location, and interactions with one another (1, 13, 15, 22, 25, 26).
Recently, mutant mice with targeted disruption of all three mPer genes have been generated and characterized (3, 5, 20, 27, 28). Studies of these mutant mice reveal that mPER1 and mPER2 have critical roles in the mammalian circadian clock, whereas mPER3 is dispensable for a functional clock. The molecular basis for the difference in clock-relevant functions among the mPER proteins is unknown, however.
We therefore sought to determine why mPER3 cannot sustain circadian clock function by comparing its posttranslational regulation with that of mPER1 and mPER2 in wild-type mice and in mice with various mPer mutations. In this way, the functions of each mPER protein could be inferred by correlating biochemical defects with the disruption of specific mPer gene(s). Our results show that mPER1 and mPER2 are redundant for the rhythmic, posttranslational regulation of clock proteins, including the phosphorylation program, the rhythmic assembly of clock protein complexes, and nuclear translocation. In vitro studies with chimeric proteins show that the inability of mPER3 to support circadian clock function results in part from lack of direct and stable interaction with CKIɛ. The data suggest that the CKIɛ-binding domain (CKBD) is an essential feature for mPER1 and mPER2 function in the circadian clock mechanism.
MATERIALS AND METHODS
Mouse strains and tissue collection.
The homozygous mPer single-mutant (mPer1ldc-, mPer2ldc-, and mPer3-deficient) and mPer double-mutant (mPer1/2-, mPer1/3-, and mPer2/3-deficient) mice used in the present study were described previously (3, 20). The wild-type mice were of the same SV129 genetic background. BALB/c mice were used in the experiments depicted in Fig. 1B to D, 2, 4, and 5. Mice were entrained to a 24-h light-dark cycle (12-h light and 12-h dark) and sacrificed on the first day in constant darkness. Tissues were dissected and frozen on dry ice.
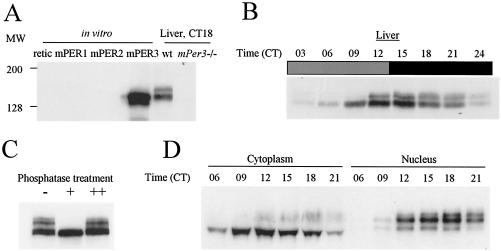
FIG. 1.
Rhythmic posttranslational modification of mPER3. (A) Characterization of antisera against mPER3. Equal amounts of in vitro translated mPER1, mPER2, mPER3, and reticulolycyte lysate were resolved by SDS-PAGE, along with liver extracts from wild-type (wt) and mPer3-deficient mice. The proteins were transferred onto nitrocellulose membrane and Western blotted. Representative Western blots with one of our strongest antisera (P3-28R) are shown. The antibody only recognized in vitro-translated mPER3 and not equal amounts of mPER1 or mPER2. Anti-mPER1 (PER1-1-R) and -mPER2 (PER2-1-R) antibodies only recognized in vitro-translated mPER1 and mPER2, respectively (data not shown). Similar sized bands were detected only in liver extracts from wild-type mice and not mPer3-deficient mice. (B) Circadian rhythms of mPER3. Liver extracts prepared from mice collected at 3-h intervals were Western blotted with P3-28R and anti-β-actin antibodies. Even loading of total protein was assured by Western blot with anti-β-actin antibody (data not shown). (C) Electrophoretic mobility shift of mPER3 is due to phosphorylation. Liver extract from CT15 was subjected to IP with anti-mPER3 antibody (P3-28GP). The immune complexes were split into three aliquots and incubated in the presence (+) or absence (−) of λPP or with both λPP and 40 mM vanadate (++). (D) mPER3 exhibits nuclear accumulation in a circadian manner. Mouse liver extracts were collected at the indicated times, fractionated into cytoplasmic and nuclear portions, and blotted for mPER3. Both cytoplasmic and nuclear fractions for each time point were derived from the same number of cells.
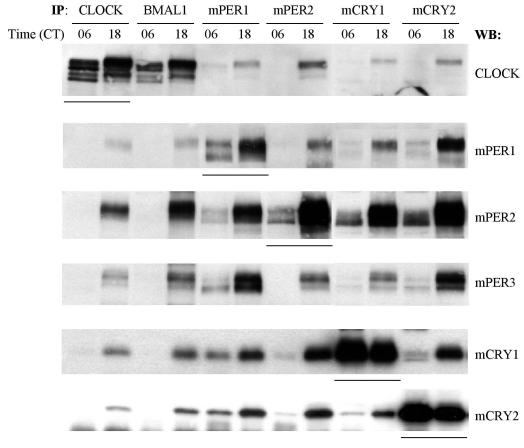
FIG. 2.
mPER3 interacts with other clock proteins in a circadian manner. Liver extracts from CT06 and CT18 were immunoprecipitated with antibodies to CLOCK, BMAL1, mPER1, mPER2, mCRY1, and mCRY2. The immune complexes were Western blotted for CLOCK, mPER1, mPER2, mPER3, mCRY1, and mCRY2. Underlined lanes indicate the primary target proteins of IP.
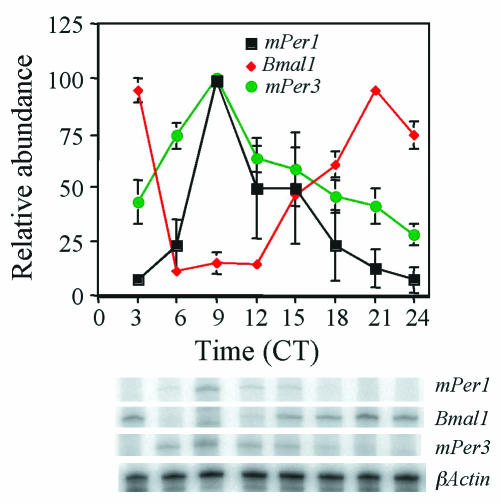
FIG. 4.
Circadian oscillation of mPer3 mRNA. mPer1, mPer3, and Bmal1 RNA rhythms in liver were determined by RNase protection assay at 3-h intervals on the first day in constant darkness (DD). Each value was normalized to β-actin and converted to the percentage of maximal value for each gene. Mean values and standard errors from three separate experiments are shown. A representative autoradiogram is shown at the bottom of the figure.
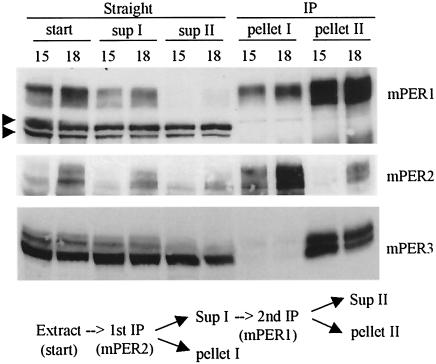
FIG. 5.
The mPER1-mPER3 complex is more abundant than the mPER2-mPER3 complex. Liver extracts from CT15 and CT18 were subjected to sequential IP reactions. Antibody to mPER2 was first added, and immune complexes were separated from supernatant. The supernatant was transferred to a new tube and subjected to a second IP with anti-mPER1 antibody. Immune complexes and supernatant from both IP reactions were analyzed by WB for mPER1, mPER2, and mPER3. Two arrowheads indicate nonspecific bands that confirm equal loading of total protein. The majority of mPER3 was copurified with mPER1, indicating that mPER1 and not mPER2 is the major binding partner for mPER3.
Recombinant plasmids.
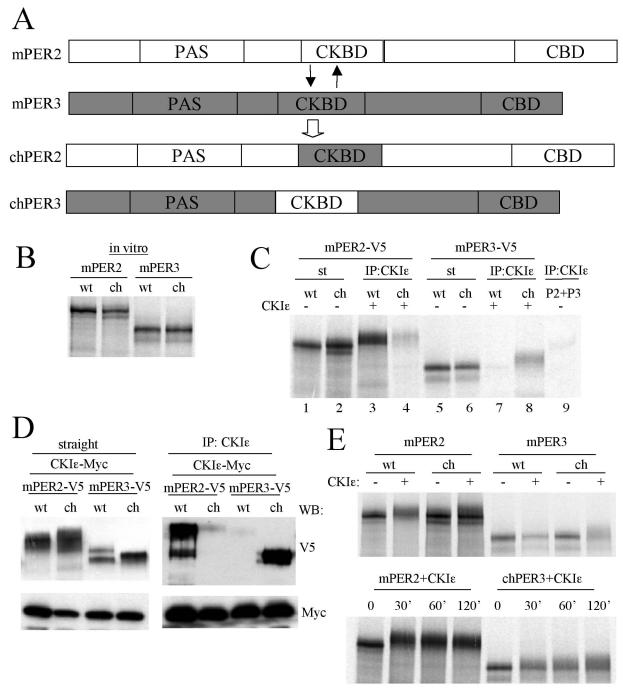
mPER1, mPER2, mPER3, mCRY1, and mCRY2 plasmids were described previously (15). To generate CKIɛ expression plasmid, the open reading frame of mouse CKIɛ (AB028736) was cloned into KpnI and XhoI sites of pcDNA3.1/myc-His(A) vector (Invitrogen). For chimeric mPER2 (chPER2) and mPER3 (chPER3) plasmids, the CKBD of mPER2 (amino acids 555 to 754) (1) was replaced with that of mPER3 (amino acids 509 to 710) (29) and vice versa. To replace the endogenous CKBD with the heterologous CKBD, NotI and XhoI were introduced into the N terminus of CKBD and the C terminus of CKBD, respectively. This resulted in the introduction of four exogenous amino acids in the chimeric PER proteins. The chimeric full-length cDNAs were cloned into KpnI and XhoI sites of pcDNA3.1/V5-His(A) vector.
Generation of anti-mPER3, anti-mCRY1, and anti-mCRY2 antibodies.
We used PCR to amplify DNA sequences that encodes amino acids 4 to 227 for mPER3 (GenBank accession no. AF050182), amino acids 486 to 606 for mCRY1 (AB000777), and amino acids 504 to 592 for mCRY2 (AB003433). The PCR product for mPER3 was cloned into BamHI and HindIII sites of PET23b, and those for mCRY1 and mCRY2 were cloned into BamHI and EcoRI sites of PET23b (Novagen). Antibodies were produced as described previously (15). Representative antisera to each clock protein were affinity purified and designated P3-28R and P3-28GP (R, raised in rats; GP, raised in guinea pigs) for mPER3, C1-R and C1-GP for mCRY1, and C2-R and C2-GP for mCRY2. Antibodies to mCRY1 and mCRY2 were tested by using in vitro translated proteins and liver extracts prepared from wild-type and mCRY-deficient mice (24). The anti-mCRY1 and mCRY2 antibodies reacted with a band present in liver extracts from wild-type mice but not in liver extracts from mCRY-deficient mice (data not shown). Similar results were obtained with anti-mCRY antibodies from Alpha Diagnostic International.
RNase protection assay.
RNase protection assays were performed as described previously. The antisense probes for Bmal1 and mPer1 were described previously (15). To make antisense probe for mPer3, we used PCR to amplify nucleotide sequences 462 to 678 of the mPer3 open reading frame (GenBank accession no. AF050182). The PCR product was cloned into EcoRI and HindIII sites of the pCR II-TOPO vector (Invitrogen). The plasmid was linearized with EcoRI and in vitro transcribed by Sp6 RNA polymerase.
In vitro translation.
In vitro translated proteins were produced in the presence of l-[35S]methionine as described previously (15).
Nuclear and cytoplasmic extracts from liver.
Subcellular fractionation of liver extracts was performed as described previously (15). Fractionation was evaluated by examining distribution of RNA polymerase II between cytoplasmic and nuclear fractions as described previously (15). More than 95% of RNA polymerase was found in nuclear fractions in all cases (data not shown).
IP and phosphatase treatment.
For frozen liver tissue, immunoprecipitation (IP) was performed as described previously (15), and the immune complexes were analyzed by Western blotting as described below. The antibodies used for IP were P3-28GP for mPER3, C1-GP for mCRY1, and C2-GP for mCRY2. Other antibodies used for IP were described previously (15). For transfected NIH 3T3 cells, IP was performed with the following modifications. NIH 3T3 cells were washed with phosphate-buffered saline and harvested 24 h posttransfection. The cells were homogenized in an Eppendorf tube with a minihomogenizer (Kontes) at 4°C in 5 volumes of extraction buffer (EB; 20 mM HEPES [pH 7.5], 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 0.05% Triton X-100, 10 μg of aprotinin/μl, 5 μg of leupeptin/μl, and 1 μg of pepstatin A/μl) and centrifuged at 12,000 × g for 10 min, and the supernatant was transferred to a new tube. Protein concentration was measured by Coomassie protein assay reagent (Pierce), and equal amounts of total protein were used for each experiment. IP was performed as described previously by using the clarified supernatant (15). Anti-hemagglutinin (HA) and V5 antibodies were purchased from Invitrogen. The antibody to immunoprecipitate CKIɛ was CKIɛ-GP (15). For in vitro-translated proteins, reticulocyte lysates containing known amounts of target proteins were mixed in different combinations as indicated in figure legends and incubated in the presence of protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 10 μg of aprotinin/μl, 5 μg of leupeptin/μl, 1 μg/μl of pepstatin A, and 1 mM dithiothreitol) at room temperature for 30 min. Subsequently, 300 μl of EB was added. The samples were precleared and subjected to IP as described previously (15). For phosphatase treatment of mPER3, liver extract was subjected to IP with anti-mPER3 antibody (P3-28GP) to obtain mPER3-containing immune complexes. The immune complexes were divided into three aliquots. Then, 200 U of lambda protein phosphatase (λPP; New England Biolabs) was added to one aliquot. To a second aliquot, 40 mM vanadate was added before the addition of λPP. No addition was made to the last aliquot. All three aliquots were incubated at room temperature for 20 min and analyzed by Western blotting as described below.
WB.
For frozen tissues, Western blotting (WB) was performed as described previously (15). To visualize mPER3, 6% sodium dodecyl sulfate (SDS)-polyacrylamide gels were used. The primary antibodies used for mPER3, mCRY1, and mCRY2 were P3-28R, C1-R, and C2-R, respectively. Other primary antibodies and a secondary antibody were as described previously (15). For transfected cells, cells were homogenized as described above, and equal amounts (∼5 μg) of total protein were analyzed by Western blotting as described previously (15).
Cell culture and transfection.
NIH 3T3 cells were grown and transfected as described previously (14). Then, 6-cm dishes were used for straight WB, and 10-cm dishes were used for IP experiments.
In vitro kinase assay.
mPERs, chPERs, and CΚIɛ were synthesized in vitro as described above. Each of the native and chimeric PERs was mixed with CKIɛ or the same volume of unprogrammed reticulocyte lysate. The molar ratio of PER to CKIɛ was 1:3. These mixtures were incubated in the presence of protease inhibitors (see above) on ice for 30 min. Subsequently, 100 μl of EB and 0.1 mM ATP were added, and these mixtures were further incubated at room temperature with gentle shaking. The reactions were terminated at 60 min after the addition of ATP for experiments depicted in Fig. 8E, upper panel. For results shown in Fig. 6E, lower panel, small aliquots from the mixtures were taken and mixed with 2× sample buffer at the indicated times after the addition of ATP. These samples were resolved by SDS-polyacrylamide gel electrophoresis (PAGE; 6% gels) and visualized by autoradiography.
FIG. 8.
CKBD of mPER is important for interaction with CKIɛ and efficient phosphorylation. (A) Diagram of CKBD swapping between mPER2 and mPER3. Amino acids 510 to 709 of mPER3 were exchanged for amino acids 551 to 750 of mPER2. (B) The apparent sizes of chPERs are the same as their wild-type counterparts. mPER2, mPER3, chPER2, and chPER3 were synthesized in vitro in the presence of l-[35S]methionine, and they were resolved by SDS-PAGE. (C) chPER3 binds to CKIɛ more efficiently than mPER3, whereas chPER2 binds to CKIɛ much less efficiently than mPER2. The PERs and CKIɛ were synthesized in vitro in the presence of l-[35S]methionine. Each of the PERs was mixed with CKIɛ at a molar ratio of 1:3. The mixtures were subjected to IP with CKIɛ antibody, and the immune complexes were resolved by SDS-PAGE and visualized by autoradiography. CKIɛ antibody immunoprecipitated >95% of input CKIɛ in all cases (data not shown). When CKIɛ was absent from the mixtures, mPER2 and mPER3 were not significantly immunoprecipitated by anti-CKIɛ antibody (last lane). Slow-migrating forms were visible for copurified PER proteins (lanes 3, 4, and 8) compared to input PERs (lanes 1, 2, and 6). (D) Results similar to those in panel C were obtained with transiently expressed proteins in NIH 3T3 cells. Native and chimeric PERs were coexpressed with CKIɛ in NIH 3T3 cells. The cell lysates were subjected to IP with anti-CKIɛ antibody, and the immune complexes were Western blotted with anti-V5 and Myc antibodies (right panel). The left panel indicates the amounts of mPERs and CKIɛ present in the cell lysates. (E) Phosphorylation of chPER3 is enhanced, but that of chPER2 is attenuated compared to their wild-type counterparts in vitro. In vitro-translated PER proteins were incubated with or without CKIɛ for 60 min. at room temperature (upper panel) or incubated with CKIɛ for the indicated times (lower panel). The mixtures were then resolved by SDS-PAGE and visualized by autoradiography. Representative autoradiograms are shown.
FIG. 6.
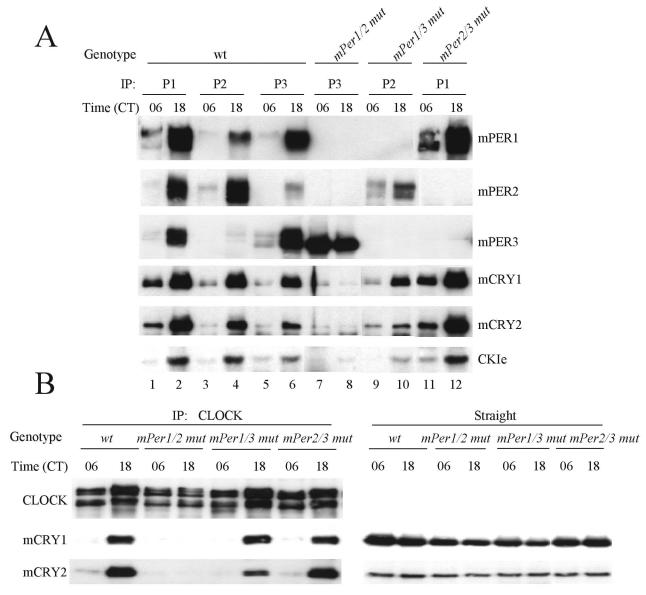
mPER3 cannot participate in rhythmic assembly of clock protein complexes. (A) mPER complexes in wild-type (wt) and mPer double-mutant mice. Liver extracts from wild-type and mPer double-mutant mice collected at CT06 and CT18 were immunoprecipitated with antibodies to mPER1, mPER2, and mPER3 (P1, P2, or P3). The immune complexes were Western blotted for mPERs, mCRYs, and CKIɛ. (B) Interaction between CLOCK-BMAL1 and mCRY is abolished in mPer1/2 double-mutant mice. Liver extracts from wild-type (wt) and mPer double-mutant mice collected at CT06 and CT18 were subjected to IP with anti-CLOCK antibody. The immune complexes were Western blotted for CLOCK, mCRY1, and mCRY2 (left panels). Straight liver extracts were Western blotted for mCRY1 and mCRY2 (right panels) to show that mCRYs are readily detectable in mice of each genotype.
RESULTS
Rhythmic phosphorylation and nuclear accumulation of mPER3.
We first examined the posttranslational regulation of mPER3 in the livers of wild-type mice. We characterized mPER3 protein in vivo by using novel antisera raised against a fragment of mPER3. One antisera (P3-28R) recognized in vitro-translated mPER3 by WB analysis but did not cross-react with equivalent amounts of mPER1 or mPER2 (Fig. 1A). Furthermore, the antibody reacted with a band whose size ranges between 135 to 150 kDa, which is present only in liver extracts from wild-type mice, but not mPer3-deficient mice.
We next examined the expression profile of mPER3 at 3-h intervals on the first day in constant darkness (DD). Like mPER1 and mPER2, mPER3 exhibited a circadian rhythm in abundance and electrophoretic mobility (Fig. 1B) (15). Rapidly migrating forms start to accumulate at circadian time 06 (CT06). Both abundance and size increase with time, peaking between CT12 and CT15. The mRNA peak of mPER3 in liver occurs at CT09, indicating a delay between mRNA peak and protein peak, as observed for the other mPERs (Fig. 4) (15). The electrophoretic mobility shift was due mainly to phosphorylation, as treatment with λPP altered the mobility of the slowly migrating forms to more rapidly migrating forms (Fig. 1C).
We showed previously that mPER1 and mPER2 translocate into the nucleus in a time-dependent manner, and hyperphosphorylated forms of mPER1 and mPER2 are detected predominantly in the nucleus (15). mPER3 also showed a striking circadian rhythm in the nuclear accumulation in liver, with hyperphosphorylated mPER3 found primarily in the nucleus (Fig. 1D).
mPER3 associates with other clock proteins in a time-dependent manner.
Our previous studies showed that seven clock proteins (CLOCK, BMAL1, mPER1, mPER2, mCRY1, mCRY2, and CKIɛ) associate as a multimeric complex in a time-dependent manner in liver (15). To test whether mPER3 is also part of this complex, we used co-IP to examine interactions of mPER3 with other clock proteins before (CT06) and during (CT18) negative feedback. Hyperphosphorylated mPER3 was copurified along with each of the seven clock proteins examined at CT18 (Fig. 2 and data not shown).
mPER1 regulates the rhythmic posttranslational modifications of mPER3.
The robust oscillations in mPER1 and mPER2 abundance and phosphorylation allow these rhythms to be used to assess the functional state of the circadian clock. Consistent with the loss of locomotor rhythmicity in mPer1/2 double-mutant mice, the double mutants lacked rhythmic posttranslational modification of mPER3 (Fig. 3A). In mPer1/3 and mPer2/3 double-mutant mice, in which molecular and behavioral rhythmicity persist for several days in DD (3), the remaining PER gene product (mPER2 and mPER1, respectively) underwent rhythmic changes in abundance and phosphorylation, as in wild-type mice, except the rhythm phases appeared advanced compared to those in wild-type mice (Fig. 3A). Thus, either mPER1 or mPER2 is sufficient for its own circadian changes in phosphorylation and abundance, whereas mPER3 is not.
FIG.3.
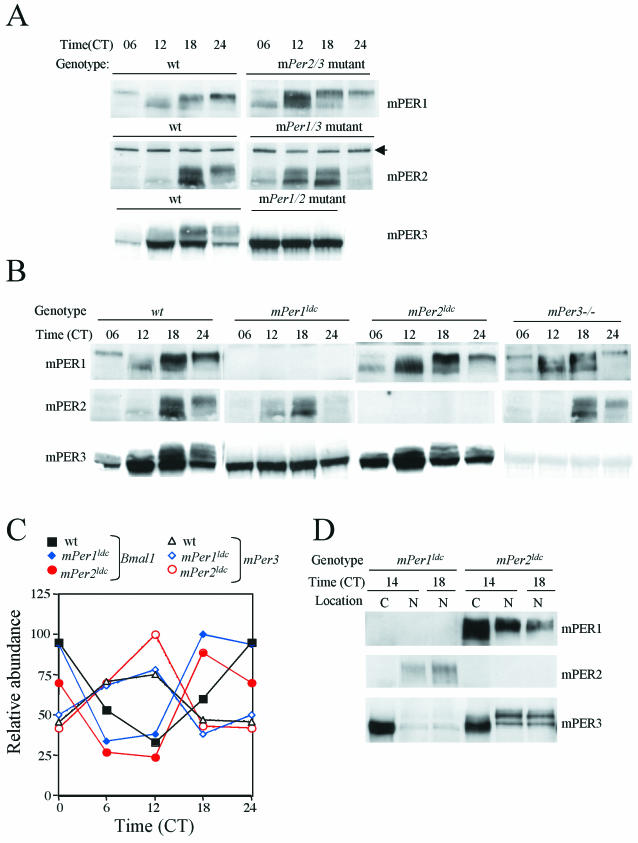
mPER3 is posttranslationally regulated by mPER1. (A) mPER profile in mPer double-mutant mice. Liver extracts from wild-type (wt), mPer1/2, mPer1/3, and mPer2/3 double-mutant mice collected at indicated times were subjected to Western blot for mPER1, mPER2, and mPER3. The black arrow indicates a nonspecific band. The results from one of two independent experiments are shown. Similar results were obtained from the other experiment (data not shown). (B) mPER profile in wild-type (wt) and mPer single-mutant mice. Liver extracts prepared from wild-type and mPer1ldc, mPer2ldc, and mPer3-deficient mice collected at the indicated times were subjected to Western blotting for mPER1, mPER2, and mPER3. (C) mPer3 RNA levels in mPer mutant mice. RNA levels were analyzed at 6-h intervals. One of two independent experiments is shown. Duplicate values varied by no more than 20% from each other. (D) mPER3 is mostly cytoplasmic in mPer1ldc mice. Liver extracts from mPer1ldc and mPer2ldc mice collected at CT14 and CT18 were fractionated and analyzed as in Fig. 1D. C and N, cytoplasmic and nuclear fractions, respectively.
The lack of rhythmic posttranslational modifications of mPER3 in mPer1/mPer2 double-mutant mice could be secondary to a broken circadian clock; that is, the posttranslational modification of mPER3 could simply be dependent upon a functional circadian clock. Alternatively, there may be a specific dependence of mPER3 on one of the other two mPER proteins for its posttranslational regulation. To dissect these possibilities, we examined the posttranslational regulation of mPER proteins in homozygous single-mutant mice. In mPer2ldc or mPer3-deficient mice, mPER1 exhibited robust oscillations in phosphorylation and abundance, as in wild-type mice (Fig. 3B). Similarly, circadian rhythms of mPER2 phosphorylation and abundance were not significantly altered in mPer1ldc or mPer3-deficient mice (Fig. 3B). mPER3 rhythms also appeared normal in mPer2ldc mice (Fig. 3B).
Remarkably, however, mPER3 did not show temporal changes in phosphorylation and abundance in mPer1ldc mutant mice (Fig. 3B). Instead, mPER3 abundance was constant, and the protein remained hypophosphorylated across the circadian cycle in the absence of mPER1. The loss of rhythmicity in mPER3 was not due to altered transcriptional regulation of mPer3 because mPer3 mRNA levels manifested robust oscillations in mPer1ldc and mPer2ldc mutant mice, as in wild-type mice (Fig. 3C). Examination of the subcellular localization of mPER3 revealed that the protein was trapped in the cytoplasm in the absence of mPER1 (Fig. 3D). Immunodepletion experiments of wild-type livers confirmed that the majority of hyperphosphorylated mPER3 is complexed with mPER1 (Fig. 5).
mPER3 cannot participate in the rhythmic assembly of clock protein complexes.
We next examined the ability of mPER3 to direct the rhythmic assembly of non-PER clock proteins by using the mPer double-mutant mice. As previously indicated (Fig. 2), mPER proteins associated with mCRY1, mCRY2, and CKIɛ more at CT18 than at CT06 in wild-type mice (Fig. 6A, lanes 1 to 6). The same interaction pattern was maintained in mPer1/3 and mPer2/3 double-mutant mice; associations between the single remaining mPER member and mCRY1, mCRY2, and CKIɛ were most apparent at CT18 (Fig.6A, lanes 9 to 12). However, the interactions between mPER3 and mCRY1, mCRY2, and kinase in mPer1/2 double-mutant mice were greatly reduced and did not appear to be rhythmic (Fig. 6A, lanes 7 and 8), suggesting that mPER3 association with these clock proteins requires mPER1 and/or mPER2. There appeared to be a decrease in mPER2 abundance in the absence of mPER1 in some experiments (Fig. 3D and 6A), but this was not a consistent finding (Fig. 3A and B).
We also examined whether the remaining mPER protein in the double-mutant mice can induce rhythmic interactions between the major transcriptional inhibitors (mCRY1 and mCRY2) and the transcriptional activators (CLOCK-BMAL1). As expected, the interactions between CLOCK and the mCRY proteins were much greater at CT18 than at CT06 in wild-type mice (Fig. 6B). The same rhythmic interactions were maintained in mPer1/3 and mPer 2/3 double-mutant mice, but these interactions were not detectable at either time in the mPer1/2 double-mutant mice (Fig. 6B). The absence of these interactions is not due to low levels of the mCRY proteins in the mutants, since the mCRYs were readily detectable in the mPer1/2 double-mutant mice and in the other mutant and wild-type mice (Fig. 6B, right panels).
Taken together, our data indicate that either mPER1 or mPER2 alone is sufficient for rhythmic posttranslational regulation of clock proteins and maintaining a functioning clock for a few circadian cycles. In contrast, mPER3 is not sufficient to support rhythmic molecular oscillations for a functioning circadian clock, implying that mPER3 is mechanistically different from mPER1 and mPER2 in clock-relevant functions.
In vitro binding assays reveal a lack of stable interaction between mPER3 and CKIɛ.
Our previous studies suggested that the interactions between mPER proteins and other clock proteins in the cytoplasm trigger nuclear translocation of mPER-containing complexes and subsequent inhibition of CLOCK-BMAL1 transcription (15). Therefore, it is possible that the inability of mPER3 to sustain the circadian clock results from the lack of interactions with CKIɛ and/or the mCRY proteins in the cytoplasm. We thus examined affinities of each mPER protein for CKIɛ and the mCRY proteins.
First, the physical interaction between each mPER protein and CKIɛ were examined in transiently transfected cells. When the kinase and each mPER protein were coexpressed in NIH 3T3 cells, CKIɛ was able to coprecipitate with mPER1 and mPER2, but not mPER3, a finding consistent with studies reported by Akashi et al. (Fig. 7A). CKIɛ-induced phosphorylation of mPER3 was enhanced when mPER1, but not mPER2, was coexpressed in NIH 3T3 cells (Fig. 7B, lane 5 versus lane 6). In agreement with our in vivo studies (Fig. 3), the data suggest that mPER3 requires mPER1 for stable interaction with CKIɛ and efficient phosphorylation.
FIG.7.
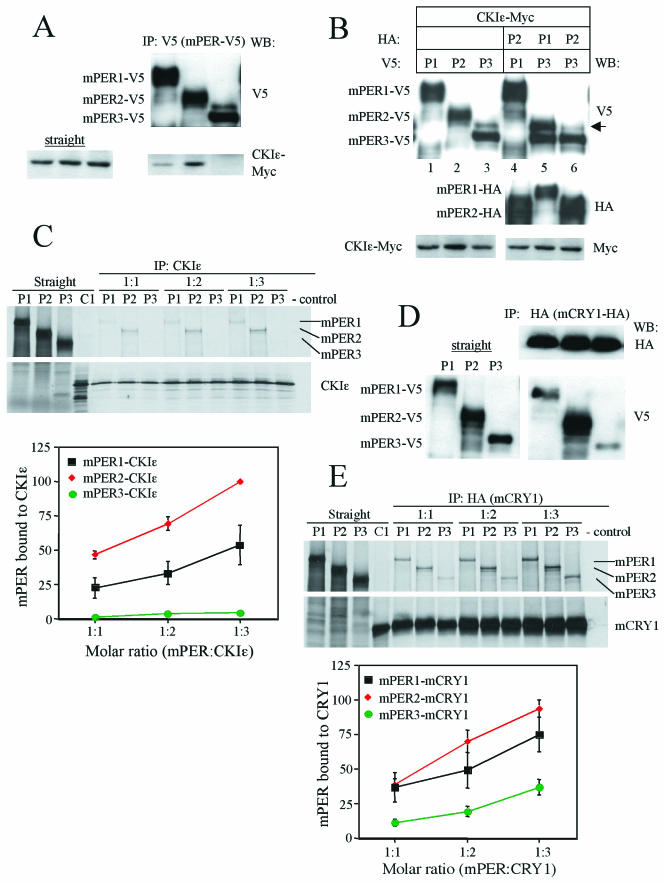
Quantitative difference of mPER protein interactions with CKIɛ and mCRY1. (A) mPER3 does not stably associate with CKIɛ in NIH 3T3 cells. NIH 3T3 cells were transfected with CKIɛ-Myc and mPER1-V5, mPER2-V5, or mPER3-V5. The cell lysates were subjected to IP with anti-V5 antibody and the immune complexes were Western blotted with anti-V5 and Myc antibodies. CKIɛ-Myc was comparably expressed in the three cell lysates (left panel). (B) CKIɛ-induced phosphorylation of mPER3 is enhanced by mPER1 but not by mPER2. CKIɛ-Myc and various combinations of mPER were expressed in NIH 3T3 cells. The cell lysates were subjected to Western blotting with anti-V5, HA, and Myc antibodies. The arrow indicates hyperphosphorylated mPER3. (C) Binding of mPER with CKIɛ. mPERs and CKIɛ were synthesized in vitro in the presence of l-[35S]methionine. Each of the mPER proteins and CKIɛ were mixed at a molar ratio of 1:1, 1:2, or 1:3. The mixtures were subjected to IP with CKIɛ antibody. The immune complexes were resolved by SDS-PAGE and visualized by autoradiography or quantified by Fuji phosphorimager. All IP reactions recovered more than 95% of input primary antigens (data not shown). A representative autoradiogram is shown (upper panel). Coimmunoprecipitated mPER was quantified from three independent experiments. Each value was converted to the percentage of the maximal value of the experiment. Mean values with standard errors are shown (lower panel). (D) NIH 3T3 cells were transfected with mCRY1-HA and mPER1-V5, mPER2-V5, or mPER3-V5. The cell lysates were subjected to IP with anti-HA antibody, and the immune complexes were Western blotted with anti-HA and V5 antibodies. The left panel shows the amounts of mPER proteins present in the three cell lysates. (E) Experiments with mPER proteins and mCRY1 were performed as in panel C to determine mPER affinity for mCRY1.
To determine the affinities of the mPER proteins for CKIɛ quantitatively, we performed binding assays with in vitro translated proteins. Each of the mPER proteins and kinase were mixed at various molar ratios and then subjected to IP with an anti-CKIɛ antibody. Subsequently, the affinity of the mPER for CKIɛ was measured by quantifying the amount of copurified mPER.
The amounts of mPER2 bound to CKIɛ were about twofold higher than those of mPER1 (Fig. 7C). Consistent with the results in Fig. 7A, we could not detect stable interaction between mPER3 and CKIɛ despite numerous attempts (Fig. 7C). These data demonstrate that mPER3 is intrinsically defective in its ability to bind to CKIɛ.
A set of similar experiments was conducted to examine interactions between the mPER and mCRY proteins. As previously reported (14), all three mPER proteins associated with mCRY1 in transiently transfected NIH 3T3 cells (Fig. 7D). The relative binding affinity of mPER2 for mCRY1 appeared to be greater than that of mPER1 or mPER3. Similar results were obtained when mCRY2 was cotransfected with each of the mPER proteins (data not shown). The affinities of the mPER proteins for mCRY1 were quantitatively determined with in vitro binding assays (Fig. 7E). The rank order of mPER affinity for mCRY1 was mPER2 > mPER1 > mPER3. At all molar ratios, the amounts of mPER3 bound to mCRY1 were two- to threefold less than those of mPER2.
Although the affinity of mPER3 for mCRY1 is lower than that of mPER1 or mPER2, the interactions between mPER3 and the mCRY proteins are readily observed in cultured cells (Fig. 7D) (14). Thus, mPER3 is not intrinsically defective in binding to the mCRY proteins. Furthermore, mPER3 is more abundant than mPER1 in liver extracts, since immunodepletion of mPER1 removed less than 50% of mPER3 from liver extracts (Fig. 5 and data not shown). Higher levels of expression would mitigate the impact of a slightly reduced affinity. We therefore conclude that the inability of mPER3 to maintain molecular or behavioral rhythms is due primarily to a deficiency in interaction with CKIɛ. The reduced affinity of mPER3 for the mCRY proteins may also contribute, but to a lesser degree.
The CKBD of mPER is essential for kinase interaction and phosphorylation.
If the direct association of mPER with CKIɛ is essential for clock-relevant functions of mPER, exchanging the CKBDs between mPER2 and mPER3 should cause predictable alterations in chimeric protein function. We thus generated the chimeric chPER2 and chPER3 proteins containing each other's CKBD (Fig. 8A). The apparent sizes of chPER2 and chPER3 were similar to their wild-type counterparts (Fig. 8B).
To determine whether the CKBD alters the functions of the hybrid proteins, we performed in vitro binding assays between the chimeric proteins and CKIɛ. As expected, the amount of chPER3 bound to CKIɛ greatly increased compared to that of native mPER3 (Fig. 8C). In contrast, the binding of chPER2 to CKIɛ was barely detectable. When the interactions were examined in transiently transfected NIH 3T3 cells, mPER2 and chPER3 were copurified with CKIɛ, whereas mPER3 and chPER2 were not (Fig. 8D). These data indicate that the CKBD determines the binding affinity of mPER proteins for CKIɛ. Even although mPER3 and chPER2 did not appreciably bind CKIɛ in cell culture, the proteins were highly phosphorylated, suggesting endogenous kinase activity not involving CKIɛ.
Next, we examined the efficiency of chPER phosphorylation by CKIɛ. Wild-type and chimeric proteins and CKIɛ were synthesized in vitro, and each of the PER proteins was mixed with CKIɛ to compare phosphorylation efficiency of the chimeric and native mPERs by CKIɛ in the absence of other interacting proteins. mPER2 and chPER3 were more readily phosphorylated by CKIɛ than were their counterparts, chPER2 and mPER3, respectively (Fig. 8E).
Taken together, these data suggest that the CKBD of mPERs not only determines kinase binding affinity but also phosphorylation efficiency. Since it has been suggested that mPER phosphorylation plays important roles in regulating interactions with other clock proteins, nuclear translocation, and degradation, our data suggest that the mPER CKBD is essential for clock-relevant functions of mPER1 and mPER2.
DISCUSSION
As in other organisms, a negative feedback loop constitutes the backbone of the molecular circadian clock in mammals (18). The mPER-mCRY complex plays an essential role in the negative feedback loop by exerting inhibitory activity on CLOCK-BMAL1-driven transcription in a time-dependent manner (14, 15). Although mCRYs play a major role in the inhibition, the timing of this inhibition seems to be regulated by mPERs (15). The present study suggests that the CKBD of an mPER protein dictates its clock-relevant functions.
Although mPer3 has been placed outside of the core feedback loop based on previous genetic studies (3, 20), we show here that mPER3 is in fact part of the transcriptional inhibitory complex and is regulated in a manner similar to the regulation of mPER1 and mPER2. Like mPER1 and mPER2, mPER3 is rhythmically phosphorylated and translocated into the nucleus, and it interacts rhythmically with other clock proteins. Among clock proteins, mPER3 is most strongly associated with mPER1. In addition, in the absence of mPER1 (mPer1ldc and mPer1/2 double-mutant mice) mPER3 phosphorylation is abolished. Thus, mPER3 is regulated by mPER1 at a posttranslational level. Consistent with these observations, phosphorylation of mPER3 by CKIɛ is enhanced in the presence of mPER1 in NIH 3T3 cells (Fig. 7B), and the nuclear translocation of mPER3 is promoted by mPER1 in NIH 3T3 cells (14). The presence of mPER3 in circadian clock protein complexes is likely mediated by interaction with mPER1.
mPER1 and mPER2 in mPer2/3 and mPer1/3 double-mutant mice, respectively, show apparently normal phosphorylation, interaction with other clock proteins, and nuclear translocation, suggesting that the single remaining mPER can support the molecular clock. The levels of the remaining mPER protein in the mutant mice were comparable to those of the mPER proteins in wild-type mice, suggesting that circadian changes of mPER abundance in liver were not significantly altered by disruption of mPer1/3 or mPer2/3 genes.
Unlike mPER1 or mPER2, mPER3 alone cannot support the molecular clock even for one cycle. mPER3 in mPer1/2 double mutant mice is apparently not phosphorylated and interacts very weakly with other clock proteins. In addition, mPER3 in the double-mutant mice is always cytoplasmic. Our data suggest that the lack of physical association of mPER3 with CKIɛ might explain the functional inadequacy of mPER3 in terms of supporting the circadian clock. In fact, CKIɛ has been implicated in the nuclear translocation of the mPER proteins (1, 22).
Our results suggest that stable interactions between CKIɛ and mPERs are important for effective phosphorylation and likely lie upstream of nuclear translocation of mPERs. If mPER3 alone cannot support the circadian clock due to the lack of stable interaction with CKIɛ, then replacement of CKBD of mPER3 with that of mPER2 should rescue the inability of mPER3 to support the clock. Our study indeed shows that chPER3 physically interacts with CKIɛ and is efficiently phosphorylated in vitro. Taken together, our data strongly suggest that the CKBD of mPERs is essential for stable interaction with CKIɛ, effective phosphorylation by CKIɛ, and possibly other clock-relevant functions, such as nuclear translocation. The importance of the CKBD for a functioning circadian clock is supported by a human genetic study showing that a mutation in the CKBD of hPER2 is associated with a severe sleeping disorder known as familial advanced sleep phase syndrome (23).
Acknowledgments
We thank Jason DeBruyne for helpful comments during the preparation of the manuscript.
This study was supported by NIH grant NS39303.
REFERENCES
- 1.Akashi, M., Y. Tsuchiya, T. Yoshino, and E. Nishida. 2002. Control of intracellular dynamics of mammalian period proteins by casein kinase Iε (CKIε) and CKIδ in cultured cells. Mol. Cell. Biol. 22:1693-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allada, R., P. Emery, J. S. Takahashi, and M. Rosbash. 2001. Stopping time: the genetics of fly and mouse circadian clocks. Annu. Rev. Neurosci. 24:1091-1119. [DOI] [PubMed] [Google Scholar]
- 3.Bae, K., X. Jin, E. S. Maywood, M. H. Hastings, S. M. Reppert, and D. R. Weaver. 2001. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30:525-536. [DOI] [PubMed] [Google Scholar]
- 4.Camacho, F., M. Cilio, Y. Guo, D. M. Virshup, K. Patel, O. Khorkova, S. Styren, B. Morse, Z. Yao, and G. A. Keesler. 2001. Human casein kinase Iδ phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett. 489:159-165. [DOI] [PubMed] [Google Scholar]
- 5.Cermakian, N., L. Monaco, M. P. Pando, A. Dierich, and P. Sassone-Corsi. 2001. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 20:3967-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunlap, J. C. 1999. Molecular bases for circadian clocks. Cell 96:271-290. [DOI] [PubMed] [Google Scholar]
- 7.Eide, E. J., E. L. Vielhaber, W. A. Hinz, and D. M. Virshup. 2002. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iɛ. J. Biol. Chem. 277:17248-17254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Etchegaray, J. P., C. Lee, P. A. Wade, and S. M. Reppert. 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421:177-182. [DOI] [PubMed] [Google Scholar]
- 9.Gekakis, N., D. Staknis, H. B. Nguyen, F. C. Davis, L. D. Wilsbacher, D. P. King, J. S. Takahashi, and C. J. Weitz. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280:1564-1569. [DOI] [PubMed] [Google Scholar]
- 10.Griffin, E. A., Jr., D. Staknis, and C. J. Weitz. 1999. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286:768-771. [DOI] [PubMed] [Google Scholar]
- 11.Harmer, S. L., S. Panda, and S. A. Kay. 2001. Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 17:215-253. [DOI] [PubMed] [Google Scholar]
- 12.Hogenesch, J. B., Y. Z. Gu, S. Jain, and C. A. Bradfield. 1998. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA 95:5474-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keesler, G. A., F. Camacho, Y. Guo, D. Virshup, C. Mondadori, and Z. Yao. 2000. Phosphorylation and destabilization of human period I clock protein by human casein kinase I epsilon. Neuroreport 11:951-955. [DOI] [PubMed] [Google Scholar]
- 14.Kume, K., M. J. Zylka, S. Sriram, L. P. Shearman, D. R. Weaver, X. Jin, E. S. Maywood, M. H. Hastings, and S. M. Reppert. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193-205. [DOI] [PubMed] [Google Scholar]
- 15.Lee, C., J. P. Etchegaray, F. R. Cagampang, A. S. Loudon, and S. M. Reppert. 2001. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 107:855-867. [DOI] [PubMed] [Google Scholar]
- 16.Lowrey, P. L., K. Shimomura, M. P. Antoch, S. Yamazaki, P. D. Zemenides, M. R. Ralph, M. Menaker, and J. S. Takahashi. 2000. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamura, H., S. Miyake, Y. Sumi, S. Yamaguchi, A. Yasui, M. Muijtjens, J. H. Hoeijmakers, and G. T. van der Horst. 1999. Photic induction of mPer1 and mPer2 in cry-deficient mice lacking a biological clock. Science 286:2531-2534. [DOI] [PubMed] [Google Scholar]
- 18.Reppert, S. M., and D. R. Weaver. 2002. Coordination of circadian timing in mammals. Nature 418:935-941. [DOI] [PubMed] [Google Scholar]
- 19.Reppert, S. M., and D. R. Weaver. 2001. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63:647-676. [DOI] [PubMed] [Google Scholar]
- 20.Shearman, L. P., X. Jin, C. Lee, S. M. Reppert, and D. R. Weaver. 2000. Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol. Cell. Biol. 20:6269-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shearman, L. P., S. Sriram, D. R. Weaver, E. S. Maywood, I. Chaves, B. Zheng, K. Kume, C. C. Lee, G. T. van der Horst, M. H. Hastings, and S. M. Reppert. 2000. Interacting molecular loops in the mammalian circadian clock. Science 288:1013-1019. [DOI] [PubMed] [Google Scholar]
- 22.Takano, A., K. Shimizu, S. Kani, R. M. Buijs, M. Okada, and K. Nagai. 2000. Cloning and characterization of rat casein kinase 1epsilon. FEBS Lett. 477:106-112. [DOI] [PubMed] [Google Scholar]
- 23.Toh, K. L., C. R. Jones, Y. He, E. J. Eide, W. A. Hinz, D. M. Virshup, L. J. Ptacek, and Y. H. Fu. 2001. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291:1040-1043. [DOI] [PubMed] [Google Scholar]
- 24.van der Horst, G. T., M. Muijtjens, K. Kobayashi, R. Takano, S. Kanno, M. Takao, J. de Wit, A. Verkerk, A. P. Eker, D. van Leenen, R. Buijs, D. Bootsma, J. H. Hoeijmakers, and A. Yasui. 1999. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398:627-630. [DOI] [PubMed] [Google Scholar]
- 25.Vielhaber, E., E. Eide, A. Rivers, Z. H. Gao, and D. M. Virshup. 2000. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I epsilon. Mol. Cell. Biol. 20:4888-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yagita, K., F. Tamanini, M. Yasuda, J. H. Hoeijmakers, G. T. van der Horst, and H. Okamura. 2002. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 21:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng, B., U. Albrecht, K. Kaasik, M. Sage, W. Lu, S. Vaishnav, Q. Li, Z. S. Sun, G. Eichele, A. Bradley, and C. C. Lee. 2001. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105:683-694. [DOI] [PubMed] [Google Scholar]
- 28.Zheng, B., D. W. Larkin, U. Albrecht, Z. S. Sun, M. Sage, G. Eichele, C. C. Lee, and A. Bradley. 1999. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400:169-173. [DOI] [PubMed] [Google Scholar]
- 29.Zylka, M. J., L. P. Shearman, D. R. Weaver, and S. M. Reppert. 1998. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20:1103-1110. [DOI] [PubMed] [Google Scholar]