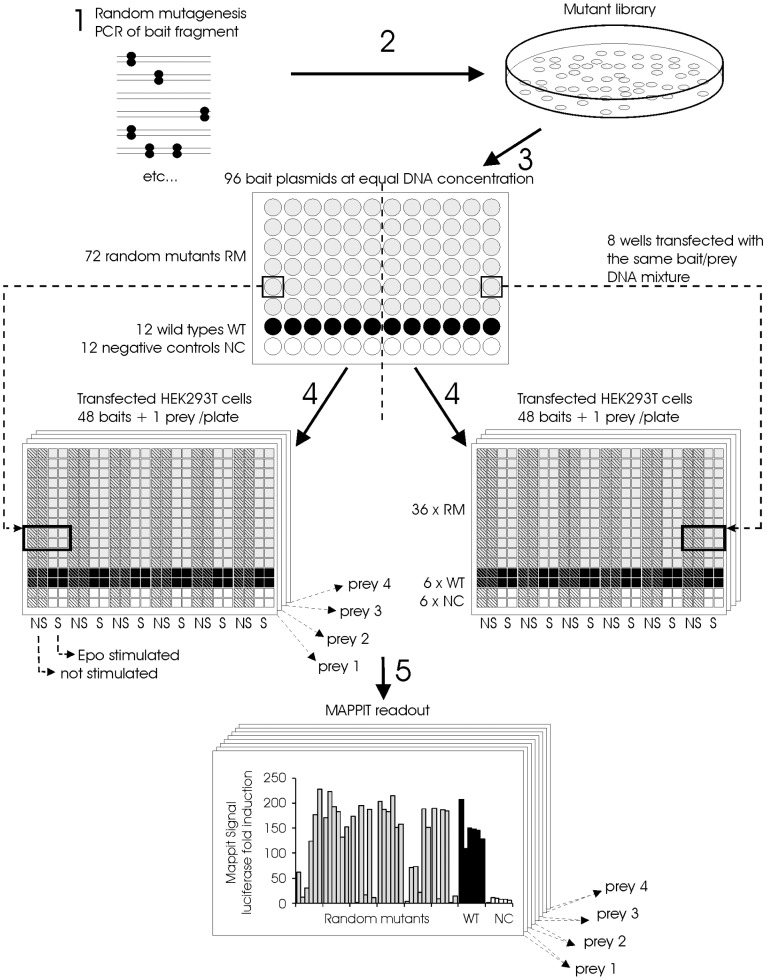
Figure 3. Method for the identification of random mutants that disrupt a protein-protein interaction based on MAPPIT.
1: A fragment of the MAPPIT bait is randomly mutated via Genemorph® II PCR. 2: The mutated PCR fragments are ligated into the MAPPIT bait vector and the resulting plasmid mutant pool is used to transform E. coli. 3: Plasmid DNA from the resulting colonies is prepared via automated 96-well DNA miniprep. Each 96-well miniprep plate contains DNA from colonies of 72 mutants, 12 wild types and 12 negative controls. The DNA concentration in all DNA samples is normalized to the same concentration. The resulting DNA is used to transfect HEK293T cells via an automated procedure using liquid handling robots. 4: Each bait is co-transfected separately with four different MAPPIT preys and the luciferase reporter. Each bait/prey mixture is used to transfect 8 wells of a 384-well plate with Hek293T cells. One 96-well MAPPIT bait miniprep plate in combination with four MAPPIT preys thus leads to 8 transfected 384 well plates. Each transfected 384 well plate contains a single MAPPIT prey in combination with 6 wild type baits, 6 negative control baits and 36 random bait mutants. 5: From each transfected bait/prey mixture, four wells are stimulated with Epo, the remaining four wells are not stimulated. After overnight Epo stimulation, the luciferase activity is determined via a luminescence reader. The MAPPIT signal is calculated by dividing the signal of the four stimulated wells by the signal of the four unstimulated wells.