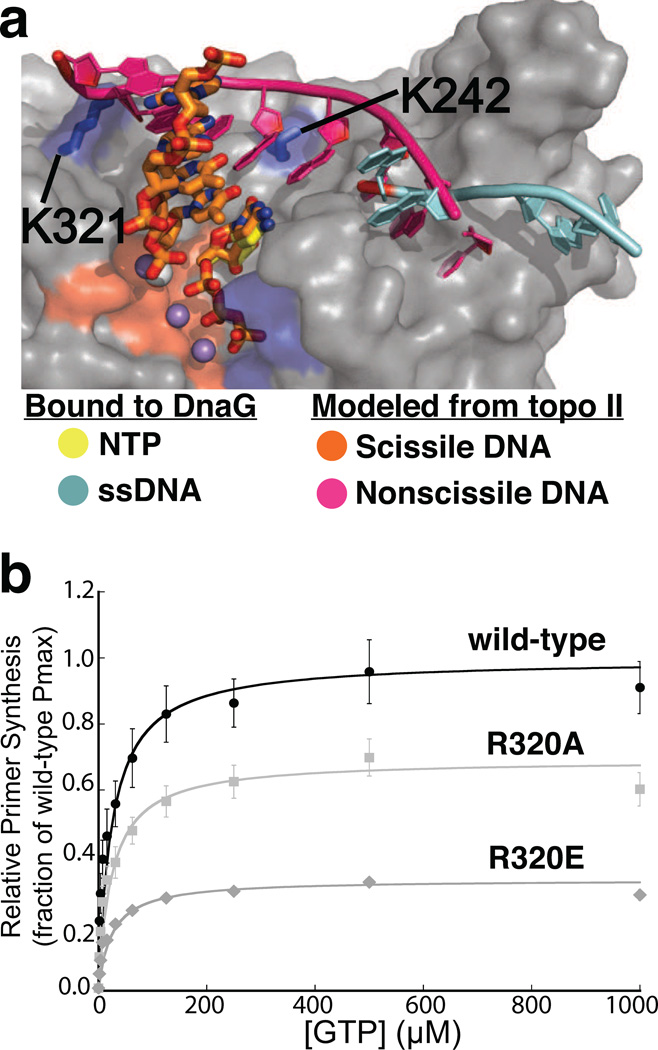
Figure 4. Congruencies between the binding sites for the complementary DNA strand in topo II DNA and SaDnaG.
a) The polarity and terminal position of the complementary DNA strand bound to the topo II TOPRIM fold lines up with the site for ssDNA binding in the DnaG RPD. A superposition of the TORPIM folds between an S. cerevisiae topo II•DNA cleavage complex and an ssDNA-bound state of the E. coli DnaG RPD (PDB ID 3B39) is shown. The scissile strand in topo II is shown as orange sticks, and its complement as a magenta cartoon. ssDNA bound to EcDnaG (cyan cartoon) is thought to mark the site of template binding (Corn and Berger, 2007; Corn et al., 2008). The protein portion of the SaDnaG RPD bound to CTP (gray surface representation) is shown, with the metal binding cluster of DnaG highlighted in red, and the basic ridge in blue. The modeling implicates residues K321 and K242 of SaDnaG (blue sticks with corresponding blue surfaces) as possibly playing a role in binding a primer•template product.
b) Arg320 of EcDnaG (corresponding to Lys321 in SaDnaG) is required for de novo primer synthesis. Helicase-stimulated, GTP-dependent primer synthesis was assayed for the wild-type enzyme (black), Arg320Ala (light gray) and Arg320Glu (gray) mutant enzymes in a fluorometric de novo primer synthesis assay. Curves were fit as described (Methods), yielding the parameters listed in Table S1; error bars represent ±SEM (Standard Error of the Mean).
See also Figure S4.