Abstract
Antisense oligonucleotides (AOs) are currently the most promising therapeutic intervention for Duchenne muscular dystrophy (DMD). AOs modulate dystrophin pre-mRNA splicing, thereby specifically restoring the dystrophin reading frame and generating a truncated but semifunctional dystrophin protein. Challenges in the development of this approach are the relatively poor systemic AO delivery and inefficient dystrophin correction in affected non-skeletal muscle tissues, including the heart. We have previously reported impressive heart activity including high-splicing efficiency and dystrophin restoration following a single administration of an arginine-rich cell-penetrating peptide (CPPs) conjugated to a phosphorodiamidate morpholino oligonucleotide (PMO): Pip5e-PMO. However, the mechanisms underlying this activity are poorly understood. Here, we report studies involving single dose administration (12.5 mg/kg) of derivatives of Pip5e-PMO, consecutively assigned as Pip6-PMOs. These peptide-PMOs comprise alterations to the central hydrophobic core of the Pip5e peptide and illustrate that certain changes to the peptide sequence improves its activity; however, partial deletions within the hydrophobic core abolish its efficiency. Our data indicate that the hydrophobic core of the Pip sequences is critical for PMO delivery to the heart and that specific modifications to this region can enhance activity further. The results have implications for therapeutic PMO development for DMD.
Keywords: antisense oligonucleotide, cardiac muscle, Duchenne muscular dystrophy, phosphorodiamidate morpholino oligomer, PMO internalizing peptide
Introduction
Duchenne muscular dystrophy (DMD) is a severe muscle wasting disorder caused by a disruption of the dystrophin mRNA reading frame resulting in an out-of-frame transcript and a non-functional dystrophin protein.1 In the past decade, a number of new treatments for DMD have been investigated, of which antisense oligonucleotide (AO)-mediated splice correction is one of the most promising approaches.2,3,4 AOs modulate dystrophin pre-mRNA splicing, by specifically restoring the reading frame of the dystrophin gene via exon skipping, and therefore generate truncated but semi-functional dystrophin protein isoforms.
In vivo studies in the DMD mouse model, mdx, have shown that systemic delivery of naked phosphorodiamidate morpholino oligomers (PMO)5 and 2′-O-methyl (2′OMe)6 AOs are not capable of restoring significant dystrophin protein in cardiac muscle. Notably, even direct intra-cardiac injections of naked AOs resulted in very low exon skipping.7 And although clinical trials with 2′OMe8,9 and PMO10,11 chemistries have shown great promise, there is still a need for further optimization to improve AO delivery to all skeletal muscles and to the heart. This is critical as respiratory complications12 and cardiac dysfunction13 are the major causes of premature death in DMD patients. In particular the cardiomyopathy becomes clinically apparent at ~10 years of age, and is exhibited in all DMD patients by the age of 18.13,14 While some studies have shown that the restoration of dystrophin in respiratory muscles can improve cardiac function in the absence of restored dystrophin protein in the heart,15,16 there is concern that any improvement in skeletal muscle function in the absence of cardiac correction may worsen the cardiac disease progression due to increases in cardiac work load.17,18,19 It is therefore highly desirable that future therapies endeavor to restore dystrophin in cardiac as well as in skeletal muscles.
Cell-penetrating peptides (CPPs), which may be readily conjugated to charge neutral AOs such as PMO, have shown potential for improved systemic delivery. CPPs that contain cationic amino acids, particularly multiple arginines, are highly effective in enhancing AO delivery due to their unique ability to deliver associated cargoes across the plasma and endosomal membranes.20 Various arginine-rich peptides have been found to be particularly effective for delivery of such charge neutral AOs,21,22,23 leading to systemic dystrophin production. However, dystrophin protein restoration in heart has typically required repeated24 and/or very high dose administrations.25
A derivative of the CPP Penetratin, originally derived from the homeobox peptide of Drosophila Antennapedia,26 has also been investigated, namely R6-Penetratin which contains six additional arginines.27 Employing R6-Penetratin as the lead peptide, a series of peptide nucleic acids/PMO internalization peptides (Pips) were derived that were found to be much more stable to serum proteolysis.28 Two such Pip peptides, Pip2a and Pip2b, conjugated to a dystrophin exon 23-specific (peptide nucleic acids) AO, were shown to be capable of inducing strong exon skipping and dystrophin positive fibres following intramuscular injection into the tibialis anterior (TA) muscle of the mdx mouse. Further optimization of this peptide series was carried out as conjugates to PMO, and Pip5e-PMO was identified as the most efficient peptide-PMO conjugate capable of inducing high levels of exon skipping and dystrophin restoration body wide, including in the heart, following a single dose intravenous administration.29 The Pip5e structure comprises a hydrophobic core region flanked on each side by arginine-rich domains containing aminohexanoyl (X) and β-alanine (B) spacers. By analogy with the previous arginine-rich B peptide,22 it was thought that the high arginine content of Pip5e contributed to overall delivery efficiency into all muscle tissues, whereas the hydrophobic region might be important for heart muscle delivery.
We now report the results of a series of mutations to the hydrophobic core region of the Pip5e peptide, where this central core region amino acid sequence is reversed, scrambled, or partially deleted. These changes affect the levels of exon skipping and dystrophin restoration in multiple muscle groups, including the heart, following a single, low dose intravenous injection of the corresponding Pip6-PMO conjugates. The results show that a core length of 5 amino acids (5-aa) appears to be essential for heart dystrophin production, since reductions in core length reduced cardiac activity. Unexpectedly, an arginine residue was tolerated in one position of the hydrophobic core, but two arginine residues were not tolerated, nor an arginine in a different position. Surprisingly, skeletal dystrophin production was also reduced in these two latter cases.
Results
Development of the Pip6 CPP series
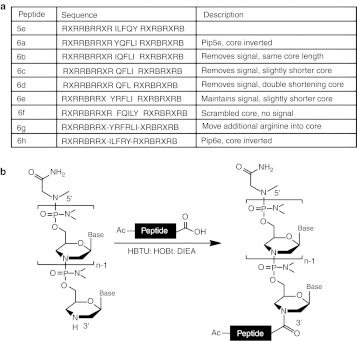
Our previous lead Pip series CPP, Pip5e,29 contains two arginine-rich flanking regions and a central hydrophobic core. To further probe the composition requirements of the hydrophobic core for maintenance of good heart dystrophin production, we synthesized a range of Pip5e derivative peptides (Pip6 a-f) (Figure 1a) where mutations were made only to the hydrophobic core region, for example scrambled and partially deleted core region peptides. All peptides contained the same number of arginine residues (10) in the flanking sequences as in Pip5e, with the exception of Pip6e. These peptides were conjugated to a 25-mer PMO complementary to dystrophin exon 23,30,31 previously validated for exon skipping in mdx mice. In contrast to the method of conjugation to the 5′ end of PMO that we utilised previously,29 Pip6-PMO conjugates were prepared by conjugation of the 3′ end of the PMO to the C-terminal carboxylic acid moiety of the Pip peptide (Figure 1b). We reported that there was no significant difference between the in vivo dystrophin production or exon skipping activity for Pip5e-PMO conjugated to the 3′ end of the PMO or to the 5′ end and therefore chose to utilise 3′ end conjugation for these experiments.32
Figure 1.
Sequences and chemical conjugation method for Pip5e-PMO derivatives. (a) List of names and sequences including rationale for synthesis of the peptides used in this study, Pip6a-h. (b) Method of conjugation of peptide to phosphorodiamidate morpholino oligonucleotide (PMO) antisense oligonucleotide (AO). Peptides were conjugated to PMO through an amide linkage at the 3′ end of the phosphorodiamidate morpholino oligonucleotide (PMO), followed by purification by high-performance liquid chromatography (HPLC) and analyzed by MALDI-TOF MS (for HPLC chromatogram and MALDI-TOF data, see Supplementary Figure S1). DIEA, diisopropylethylamine; HBTU, 2-(IH-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate; HOBt, 1-hydroxybenzotriazole monohydrate.
In vitro screening of Pip6-PMO compounds
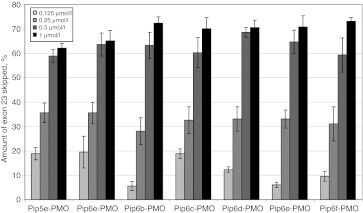
The exon skipping potential of Pip6-PMO conjugates was evaluated in differentiated mouse H2K mdx myotubes in the absence of any transfection agent (Figure 2) at concentrations ranging from 0.125 to 1 µmol/l. This showed that exon skipping activity in cultured muscle cells was very similar for all these constructs, including Pip5e-PMO. These results differ from the previous Pip5 series,29 where the flanking arginine-rich sequences mostly contained a fixed number of arginine residues (10) but where spacings were varied through alternative placement of aminohexanoyl and β-alanine units. This resulted in small variations in exon skipping activity that correlated well with in vivo activity. In the case of Pip6 sequences, the flanking arginine-rich sequences are identical (with the exception of Pip6e, which is identical except for one arginine immediately preceding the core which is displaced into the second position of the core). The results demonstrate that cellular exon skipping activity does not depend on the sequence or length of the hydrophobic core. Note that we have previously shown that major changes in in vitro exon skipping activities are correlated instead with the total numbers of arginine residues.33
Figure 2.
Exon skipping activity of Pip6-PMOs in differentiated mouse H2K mdx myotubes. H2K mdx myotubes were incubated with peptide-PMO conjugates at concentrations ranging between 0.125 and 1 µmol/l without the use of transfection reagent. The products of nested reverse transcription-PCR (RT-PCR) were examined by electrophoresis on a 2% agarose gel. Exon skipping activity is presented as the percentage of Δ23 skipping as calculated by densitometry. PMO, phosphorodiamidate morpholino oligonucleotide.
Alterations to the Pip5e hydrophobic sequence improve splicing activity in vivo
Given the potency of Pip5e-PMO in heart tissue, the aim of altering the sequence of the hydrophobic core (whilst maintaining the 5-aa length) was to identify peptides that might be more efficient at lower doses. These modifications included inversion of the hydrophobic region (Pip6a), substitution of tyrosine by isoleucine (Pip6b), substitution of glutamine in the Pip6a sequence by displacement of the arginine immediately flanking the core in the first arginine-rich flanking region (Pip6e), and a scrambled hydrophobic core sequence (Pip6f). All Pip6 peptide-PMO conjugates were administered to mdx mice as single 12.5 mg/kg intravenous injections via the tail vein and tissues were harvested 2 weeks later and assessed for activity at both RNA and protein levels.
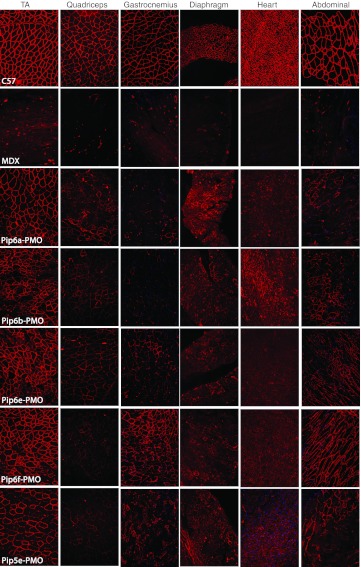
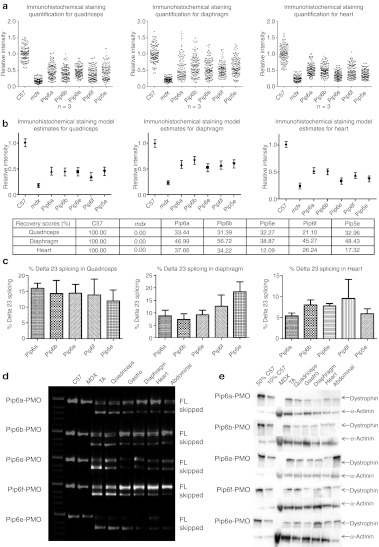
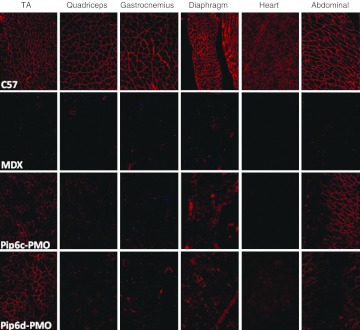
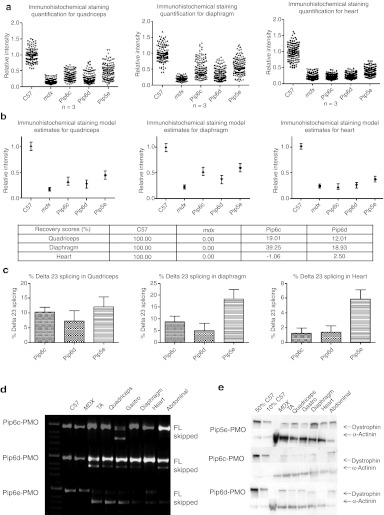
Immunohistochemical staining of dystrophin expression for all 5-aa core Pip6-PMOs revealed high levels of dystrophin production in skeletal muscles including the TA, diaphragm, and the heart (Figure 3). Immunohistochemical staining quantification (Figure 4a, b) was performed as previously described16,34 and was achieved by taking four representative frames of the dystrophin staining and correlating this with laminin staining for each section (n = 3) of the quadriceps, diaphragm and heart for each peptide-PMO treatment. Untreated mdx and treated mdx mice were normalized to C57BL10 mice. This method allows comparison of the staining intensity of dystrophin at the sarcolemma relative to laminin for each treatment group. Intensity ratios are normalized to C57BL10 samples and each region of interest at the sarcolemma (120 regions for each treatment group) is plotted on a scatter graph. The relative intensity values obtained for all four of the 5-aa core Pip6-PMO conjugates were significantly different to those of untreated mdx mice for the quadriceps and diaphragm (Figure 4b and Table 1). There were very similar dystrophin restoration levels in the quadriceps (percent recovery score—%RS—range between 21.10 and 33.44%; Figure 4b) and in the diaphragm for all treatments, with the exception of Pip6b which had a higher recovery score in the diaphragm (%RS range between 38.87 and 48.43%, Pip6b 56.72%; Figure 4b). All 5-aa core Pip6-PMO–treated mice exhibited high dystrophin intensity values in the heart with the exception of Pip6e (other Pip6-PMOs were statistically significant compared to mdx = P < 0.0001; Table 1). Pip6a- and Pip6b-PMO conjugates displayed the highest recovery scores, as observed in Figure 4b (%RS 37.66% and 34.22%, respectively) closely followed by Pip6f-PMO (%RS 26.24%) and then Pip5e-PMO (%RS 17.32%). When directly compared to Pip5e-PMO treatment, only Pip6a-PMO was significantly better in the heart (Table 2). These 5-aa core Pip6-PMOs were also shown to restore other dystrophin complex proteins, namely neuronal nitric oxide synthase, α-sarcoglycan, and β-sarcoglycan as illustrated by immunohistochemical staining in the TA muscle (Supplementary Figure S4).
Figure 3.
Immunohistochemical staining for dystrophin in C57BL10 control, mdx untreated, the 5-aa hydrophobic core Pip6-PMO-treated and Pip5e-PMO-treated mice. A single 12.5 mg/kg systemic injection of peptide-PMO was administered to mdx mice. Tissues were harvested 2 weeks later. Dystrophin immunostaining in TA, quadriceps, gastrocnemius, diaphragm, heart, and abdomen muscle groups for C57BL10, mdx-treated and mdx-untreated mice are shown. PMO, phosphorodiamidate morpholino oligonucleotide.
Figure 4.
Dystrophin splicing and protein restoration in C57BL10 control, mdx untreated, the 5-aa hydrophobic core Pip6-PMO-treated and Pip5e-PMO-treated mice following a single 12.5 mg/kg, intravenous (i.v.) injection. (a) Quantification of dystrophin immunohistochemical staining relative to control laminin counter-stain in quadriceps, diaphragm and heart muscles of C57BL10, mdx-untreated and mdx-treated mice. Relative intensity values for each region of interest (120 regions) are plotted and the model estimate average calculated (presented in b) from the repeated measures, multilevel statistical model. For statistical significance tables see Tables 1 and 2. Percentage recovery score is represented below. (c) Percentage Δ23 exon skipping as determined by quantitative real time (q-RT)-PCR in quadriceps, diaphragm, and heart muscles. (d) Representative real time (RT)-PCR images demonstrating exon skipping (skipped) in TA, quadriceps, gastrocnemius, diaphragm, heart, and abdomen muscles. The top band indicates full-length (FL) or unskipped transcript. (e) Representative western blot images for each treatment. Ten micrograms of total protein was loaded (TA, quadriceps, gastrocnemius, diaphragm, heart, and abdomen muscles) relative to 50% (5 µg protein) and 10% (1 µg) C57BL10 controls, and normalized to α-actinin loading control (for quantification see Supplementary Figure S2a). PMO, phosphorodiamidate morpholino oligonucleotide.
Table 1. Statistical tables for quantitative immunohistochemical staining mean values for C57BL10 control, Pip6-PMO-treated mdx mice and Pip5e-PMO-treated mdx mice relative to mdx-untreated, following a single 12.5 mg/kg, i.v. injection.
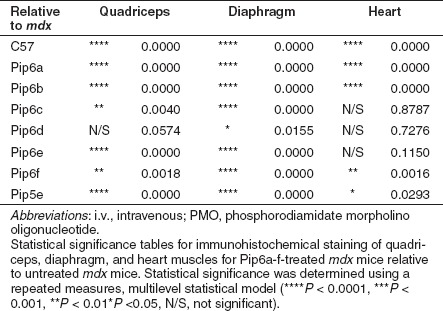
Table 2. Statistical tables for quantitative immunohistochemical staining mean values for C57BL10 control, mdx-untreated, and Pip6-PMO-treated mdx mice relative to Pip5e-PMO-treated mice, following a single 12.5 mg/kg, i.v. injection.
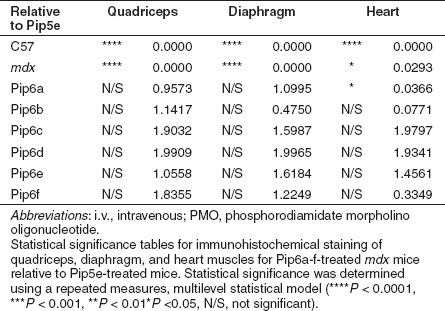
The PCR and western blot analyses exhibited similar results to the immunostaining. The reverse transcription-PCR (RT-PCR) representative images (Figure 4d) illustrate high exon skipping efficiency in all tissues analyzed. This is better shown by quantitative real-time PCR (qRT-PCR) results for the quadriceps, diaphragm and heart (Figure 4c). The delta 23 transcript is normalized against “total dystrophin” for each muscle group (n = 3). Quantification of this data revealed similar levels of Δ23 skipping in the quadriceps of all 5-aa Pip6-PMO–treated mice. The data trends suggest that Pip6f- and Pip5e-PMO show the highest exon skipping in the diaphragm, and Pip6f-PMO the highest in heart (for splicing mean values, see Supplementary Figure S2a). Western blots (Figure 4e) were performed on the tissues of each mouse and were quantified against a 50 and 10% C57BL10 control. These results were averaged and are presented in Supplementary Figure S2b. Pip6a-, Pip6b-, Pip6e-, and Pip6f-PMO conjugates exhibited the highest dystrophin protein restoration in the TA and quadriceps muscles. The levels of dystrophin restoration in the diaphragm were uniform across all of these treatments, whereas in the case of the heart, Pip6b- and Pip6f-PMO conjugates showed the highest dystrophin restoration.
Protein restoration as measured by immunohistochemical staining is consistently higher than protein restoration calculated by western blot analysis. These differences may be attributed to the differing “housing proteins” used i.e., dystrophin restoration quantified by immunohistochemical staining is normalized against laminin, whereas western blot analysis uses α-actinin for normalisation. Quantification of western blots has only recently been reported for dystrophin and currently uses chemiluminescence methods. It may therefore be judicious to give greater weight to the trends in dystrophin protein levels revealed by western blot rather than to the absolute values. Therefore, considering the results overall, mdx mice treated with each of the four 5-aa core Pip6-PMOs (Pip6a-, Pip6b-, Pip6e-, and Pip6f-PMO) appear to demonstrate improved dystrophin production and exon skipping in TA, quadriceps, and heart muscles compared with the previous lead candidate, Pip5e-PMO.
In addition, these 5-aa core Pip6-PMOs do not exhibit evidence of toxicity, as assessed by plasma levels of relevant toxicity biomarkers, alanine aminotransferase, aspartate aminotransferase, and creatine kinase (see Supplementary Figure S5a). Blood urea nitrogen and creatinine levels were similar to untreated mdx levels (see Supplementary Figure S5b). All Pip6-PMO treatment groups exhibit similar biomarker levels to untreated C57BL10 controls.
Partial deletions of the hydrophobic core of Pip6-PMO conjugates abolish heart dystrophin production
In addition to the need to identify Pip-PMOs with high efficiency and cardiac delivery, it was a further aim to better define those elements of the hydrophobic core of Pip peptides that are important for heart delivery. To this end, Pip peptides containing partial deletions of the hydrophobic core by 1 aa (removal of tyrosine; Pip6c) and by 2 aa (removal of isoleucine and tyrosine; Pip6d) were synthesised as PMO conjugates (Figure 1a).
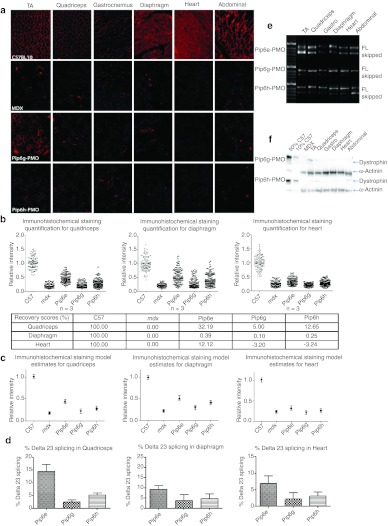
Following treatment of mdx mice, immunohistochemical staining was performed and this revealed some dystrophin expression in skeletal muscles such as the TA and diaphragm for both these deletion Pip6-PMOs (Figure 5). Quantification of the immunohistochemical staining revealed the lowest dystrophin restoration in the quadriceps with Pip6d-treatment, closely followed by Pip6c-PMO when compared to the other Pip6-PMOs (Figure 6a,b and Table 1). Similarly, Pip6d-PMO displayed the lowest dystrophin restoration in the diaphragm. The recovery scores for Pip6c- and Pip6d-PMO conjugates in the heart were very low, indicating their poor efficiency (Pip6c %RS –1.06% and Pip6d 2.50%; Figure 6b).
Figure 5.
Immunohistochemical staining for dystrophin in C57BL10 control, mdx untreated and the shortened hydrophobic core Pip6-PMO-treated mice (Pip6c and Pip6d). A single 12.5mg/kg systemic injection of PPMO was administered to mdx mice. Tissues were harvested 2 weeks later. Dystrophin immunostaining in TA, quadriceps, gastrocnemius, diaphragm, heart and abdomen muscle groups for C57BL10, mdx-treated and mdx-untreated mice are shown. PMO, phosphorodiamidate morpholino oligonucleotide.
Figure 6.
Dystrophin splicing and protein restoration in C57BL10 control, mdx untreated and the shortened hydrophobic core Pip6-PMO-treated mice (Pip6c and Pip6d) compared to Pip5e-PMO following a single 12.5 mg/kg, intravenous (i.v.) injection. (a) Quantification of dystrophin immunohistochemical staining relative to control laminin counter-stain in quadriceps, diaphragm and heart muscles of C57BL10, mdx-untreated and mdx-treated mice. Relative intensity values for each region of interest (120 regions) are plotted and the model estimate average calculated (presented in b) from the repeated measures, multilevel statistical model. For statistical significance tables, see Tables 1 and 2. Percentage recovery score is represented below. (c) Percentage Δ23 exon skipping as determined by quantitative real time (q-RT)-PCR in quadriceps, diaphragm, and heart muscles. (d) Representative real time (RT)-PCR images demonstrating exon skipping (skipped) in TA, quadriceps, gastrocnemius, diaphragm, heart, and abdomen muscles. The top band indicates full-length (FL) or unskipped transcript. (e) Representative western blot images for each treatment. Ten micrograms of total protein was loaded (TA, quadriceps, gastrocnemius, diaphragm, heart, and abdomen muscles) relative to 50% (5 µg protein) and 10% (1 µg) C57BL10 controls, and normalized to α-actinin loading control (for quantification see Supplementary Figure S2a). PMO, phosphorodiamidate morpholino oligonucleotide.
These results are corroborated by the PCR and western blot analyses. The RT-PCR representative images (Figure 6d) and the qRT-PCR exon skipping results (Figure 6c) both indicate reduced exon skipping in mdx mice treated with Pip6c- and Pip6d-PMO conjugates in quadriceps and diaphragm and negligible exon skipping in the heart. Western blot analysis revealed inefficient dystrophin protein production in the TA and quadriceps muscles and negligible dystrophin restoration in the heart (Figure 6e and Supplementary Figure S2b).
These results show that the length of the hydrophobic core is crucial not only for good heart dystrophin production but also for activity in some other muscle groups. Therefore, the arginine content of the CPP alone is not the sole predictor of dystrophin production and exon skipping efficiency for this class of peptides.
Altering the position of arginine in the hydrophobic core or adding a second arginine is detrimental to dystrophin production
The repositioning of an arginine from a flanking region into the core was unexpectedly well-tolerated (Pip6e-PMO) as it gave similar results to the Pip5e-PMO conjugate. Two further Pip6-PMO conjugates were thus synthesized (Figure 1a) as derivatives of Pip6e-PMO. Pip6g-PMO contained a second arginine residue, which was moved from the second flanking region into the central hydrophobic core, and Pip6h-PMO contains an inversion of the Pip6e hydrophobic region, such that the single arginine location is altered within the core.
Surprisingly, these changes to the hydrophobic core resulted in further reductions in dystrophin expression not only in heart but also in all other tissues, as observed in the immunohistochemical staining (Figure 7a) and in the quantifications thereof (Figure 7b,c). The immunohistochemical staining representative images revealed very few dystrophin positive fibers in all tissues with the exception of the TA and quadriceps (Figure 7a). With reference to the quantifications, Pip6g-PMO was not significantly different to untreated mdx in the quadriceps or diaphragm (Supplementary Figure S3a). Both Pip6e-PMO derivatives were not significantly different to untreated mdx in heart muscles, illustrating the general inefficiency of these two peptides. Similarly, these Pip6e-PMO derivatives showed reduced efficiency in exon skipping, as illustrated in representative RT-PCR images (Figure 7e) and in qRT-PCR analyses (Figure 7d and Supplementary Figure S3c) in all tissues. Western blots revealed negligible dystrophin protein restoration (Figure 7f and Supplementary Figure S3d) in all tissues with the exception of the TA muscle. These data show that an increase in the number of arginines or alteration in the location of the single arginine in the hydrophobic region of Pip6e are detrimental to both heart as well as skeletal muscle dystrophin production.
Figure 7.
Dystrophin splicing and protein restoration in C57BL10 control, mdx untreated and the Pip6e-PMO derivatives, Pip6g and Pip6h, following a single 12.5 mg/kg, intravenous (i.v.) injection. (a) Immunohistochemical staining for dystrophin in C57BL10 control, mdx untreated and Pip6g- and Pip6h-PMO-treated mice. Dystrophin immunostaining in TA, quadriceps, gastrocnemius, diaphragm, heart, and abdomen muscle groups for C57BL10, mdx-untreated and mdx-treated mice are shown. (b) Quantification of dystrophin immunohistochemical staining relative to laminin counter-stain in quadriceps, diaphragm, and heart muscles of C57BL10, mdx-untreated and mdx-treated mice. Relative intensity values for each region of interest (120 regions) are plotted and the model estimate averages calculated (presented in c) from the repeated measures, multilevel statistical model. For statistical significance tables see Supplementary Figure S3a,b. Percentage recovery score is represented below. (d) Percentage Δ23 exon skipping as determined by quantitative real time (q-RT)-PCR in quadriceps, diaphragm and heart muscles. (e) Representative real-time (RT)-PCR images demonstrating exon skipping (skipped) in TA, quadriceps, gastrocnemius, diaphragm, heart, and abdomen muscles. The top band indicates full-length (FL) or unskipped transcript. (f) Representative western blot images for each treatment. Ten micrograms of total protein was loaded (TA, quadriceps, gastrocnemius, diaphragm, heart, and abdomen muscles) relative to 50% (5 µg protein) and 10% (1 µg) C57BL10 controls, and normalized to α-actinin loading protein (for quantification, see Supplementary Figure S3c). PMO, phosphorodiamidate morpholino oligonucleotide.
Discussion
The most promising therapy to date for the severely debilitating neuromuscular disorder DMD is treatment with AOs, which restores the reading frame of the dystrophin pre-mRNA by exon skipping. Two AOs, a PMO10,11 and a 2′OMe oligonucleotide,8,9 are currently in clinical trials and the early promising results have increased hope for DMD patients. However, studies involving the administration of very high doses of naked PMO into mdx mice have shown only partial restoration of dystrophin in body-wide skeletal muscles and negligible correction in heart.16,35 The necessity to correct dystrophin in the heart is ever more apparent following studies whereby the correction of the skeletal phenotype resulted in an increase in the cardiac workload and thus further progression of the cardiomyopathy.17,18 The discovery that CPP-conjugated PMOs can achieve much more effective dystrophin correction in mdx mice than naked PMOs has brought renewed promise for enhanced AO efficacy by improving cellular and in vivo delivery.
We previously reported a promising peptide-PMO candidate, Pip5e-PMO, capable of restoring dystrophin protein to high levels in all muscle types, including heart, following a single 25 mg/kg administration.29 In addition to arginine-rich sequences, Pip peptides contain a 5-aa hydrophobic section not present in the previous B-peptide lead,26 which seemed likely to be responsible for the improved heart activity. The Pip6 series was developed as derivatives of Pip5e-PMO in an attempt to cast light on aspects of the hydrophobic core required for heart dystrophin production and also to identify even more active Pip-PMO conjugates. Our study using a moderate, single dose administration regimen has produced some interesting and sometimes surprising results.
A key finding is that maintenance of the 5-aa length of the hydrophobic core region is imperative for good heart dystrophin production. One might imagine that diminished efficiency of dystrophin restoration in the heart for Pip6c-PMO and Pip6d-PMO, with sequential amino acid deletions in the core, might be correlated with the resultant lower hydrophobicity and hence a reduced capacity to enter the cell.36 However, the in vitro results would suggest that all of these constructs are capable of entering the cells as they are all fully capable of exon skipping in muscle cells (Figure 2). Thus, the length of the 5-aa hydrophobic core must affect a different parameter essential for in vivo heart delivery. Enhanced uptake into whole heart slices of fluorescently labeled Pip5e-PMO, compared to B-PMO, suggested instead that crossing of another barrier (for example, the endothelial lining to the heart) is improved.30 Further heart studies are continuing with a Pip6-PMO that may help to address this issue. More surprising perhaps is that for Pip6c-PMO and Pip6d-PMO there was also some loss of dystrophin production in other muscle types. This suggests that the hydrophobic/cationic balance and/or the precise spacings of hydrophobic and cationic residues in the CPP impose more subtle effects on in vivo delivery parameters.
Another clear conclusion arising from the Pip6-PMO analogues is that a specific order of hydrophobic residues within the hydrophobic core is less important at maintaining the heart dystrophin production, since an inverted sequence (Pip6a), a single substitution of an equally hydrophobic residue (Pip6b), and a scrambled sequence (Pip6f) were at least as active as Pip5e-PMO, and more efficient in heart and some muscle groups (Figure 4 and Supplementary Figure S2). These results provide evidence that the hydrophobic core of Pip peptides is unlikely to contain a particular amino acid sequence that recognizes a specific receptor in a membrane barrier required to penetrate heart tissue, but instead the core acts as a hydrophobic spacer of some kind.
Most surprising however is that Pip6e-PMO did induce some dystrophin splicing and protein restoration in heart muscle as indicated by the western and qRT-PCR results (note: not significantly different in immunohistochemical staining quantification). In the Pip-6e peptide, one arginine residue is moved into the hydrophobic core, which also results in alignment of a hydrophobic X residue adjacent to the core (X-YRFLI). One might have expected heart dystrophin production to have been completely lost in this conjugate, since a cationic amino acid (arginine) is now included in the core. By contrast, such heart activity was lost for Pip6h-PMO (X-ILFRY core) and the double arginine core conjugate Pip6g-PMO (X-YRFRLI-X core). Unexpectedly, dystrophin production was also lost in quadriceps and diaphragm for both Pip6g- and Pip6h-PMO. The unanticipated inconsistencies within the activity results for Pip6e, Pip6g, and Pip6h, and the losses of activities for Pip6c- and Pip6d-PMO, are perhaps best explained by the realization that precise spacings of the arginine residue within the Pip peptides with respect to both the outer hydrophobic amino acid spacers (X and B) and the inner hydrophobic core residues may drastically alter the pharmacological properties of each conjugate. This might occur not only through alteration in cationic/hydrophobic balance but alternatively due to secondary or tertiary structure changes of the Pip-PMOs, which could in turn affect serum protein binding or another parameter that alters the circulatory half-life, or which could affect the ability to traverse barriers required to penetrate muscle tissues. Such more subtle effects will require a more wide-ranging pharmacological and biophysical study, upon which we are currently embarking.
For a complete understanding of the role of the hydrophobic core within Pip peptides, one would ideally wish to study the activities, pharmacology and biophysical parameters of a much larger range of sequence-variant Pip-PMO conjugates. The need for multi-mg synthesis of each conjugate as well as the availability of a large number of mdx mice has necessarily limited the scope of the current study. It is gratifying that even with the relatively small numbers of Pip6 conjugates tested, some interesting results have emerged and that several conjugates (Pip6a, Pip6b, and Pip6f-PMOs) have shown promising dystrophin production activities even beyond that of the previous candidate, Pip5e-PMO. Interestingly, analysis of serum samples from one of the 5-aa Pip6-PMO treatments, Pip6e-PMO, showed partial normalization of three miRNAs (miR-1, miR-133a, and miR-206) to near wild type levels following a single, 12.5 mg/kg administration (Roberts et al., related manuscript37). This is greatly promising for the Pip6-PMOs, as it would not be considered the optimal peptide yet still demonstrated the significant therapeutic effect of this group of peptides. These new leads provide a good basis for identification of a Pip-PMO candidate suitable for detailed physiological studies of muscle and heart function, as well as thorough toxicity profiling including dose escalation studies, in anticipation that one such Pip-PMO will proceed to clinical trial.
Materials and Methods
Synthesis of peptide-PMO conjugates. Peptides were synthesized by standard Fmoc chemistry and purified by high-performance liquid chromatography. The PMO sequence (5′-GGCCAAACCTCGGCTTACCTGAAAT-3′) was purchased from Gene Tools LLC (Corvallis, OR). Peptides were conjugated to PMO through an amide linkage at the 3′ end of the PMO, followed by purification by high-performance liquid chromatography and analyzed by MALDI-TOF MS as previously described in preliminary communication.32 Full details of synthesis including improvements to the experimental procedures are described in detail in the Supplementary Materials and Methods. Peptide-PMO conjugates were dissolved in sterile water and filtered through a 0.22-µm cellulose acetate membrane before use.
Conjugates of PMO of Pip6a, Pip6b, Pip6e, and Pip6f were found to be predominantly stable and of similar stability to Pip5e-PMO in 100% serum for 2 hours at 37 °C, as seen by high-performance liquid chromatography and MALDI-TOF mass spectral analysis. The conjugates all showed similar degradation patterns, and intact conjugates were still observed up to 4 hours (data not shown).
In vitro assays: exon skipping in mdx mouse myotubes. H2K mdx myotubes were prepared and incubated with peptide-PMO conjugates in the absence of any transfection agent at concentrations of 0.125, 0.25, 0.5, and 1.0 µmol/l by the method described previously.29 The products of nested RT-PCR from total isolated RNA were examined by electrophoresis on a 2% agarose gel. Quantification of Δ23 transcript levels was calculated using densitometry. The MTS cell viability test (Promega, Madison, WI) showed 100% survival at the highest concentrations of peptide-PMO conjugates used in the study (data not shown).
Animals and intravenous injections. Four and a half month old to 5½-month-old mdx mice were used in these experiments (n = 3). The experiments were carried out in the Biomedical Sciences Unit, University of Oxford according to procedures authorized by the UK Home Office. Pip6-PMO conjugates were prepared in 0.9% saline solution at a final dose of 12.5 mg/kg. The 160 µl total volume was administered via the tail vein of anaesthetized mice. Two weeks later mice were sacrificed by CO2 inhalation, and muscles and other tissues harvested and snap-frozen in cooled isopentane before storage at –80 °C.
Immunohistochemistry and quantification of dystrophin expression. Transverse sections of tissue samples were cut (8-µm thick) for the examination of dystrophin expression. For dystrophin visualisation and quantification, sections were co-stained with rabbit-anti-dystrophin (Abcam, Cambridge, MA) and rat anti-laminin (Sigma, St Louis, MO), and detected by goat-anti-rabbit immunoglobulin G Alexa 594 and goat-anti-rat immunoglobulin G 488 secondary antibodies, respectively (Invitrogen, Carlsbad, CA). Images were captured using a Leica DM IRB microscope and Axiovision software (Carl Zeiss, Cambridge, UK). Quantitative immunohistochemistry was performed as previously described.16,34 A representative image for each treatment was taken. For quantification, four representative frames of the dystrophin and correlating laminin were taken for each section (n = 3) of the quadriceps, diaphragm and heart for each treatment. Using ImagePro software, 10 regions of interest were randomly placed on the laminin image which was overlaid on the corresponding dystrophin image. The minimum and maximum fluorescence intensity for 120 regions were recorded for each treatment. The intensity difference was calculated for each region to correct for background fluorescence and untreated mdx and treated mdx were normalized to C57BL10. These values were plotted on a scatter graph. The “relative intensity means” were calculated using a multilevel statistics model. Using these values, the percentage recovery score was calculated by implementing the following equation, as described on the TREAT-NMD website (http://www.treat-nmd.eu/downloads/file/sops/dmd/MDX/DMD_M.1.1_001.pdf): (dystrophin recovery of treated mdx mice-dystrophin recovery of untreated mdx mice)/(dystrophin recovery of C57BL10 mice-dystrophin recovery of untreated mdx mice). Staining of dystrophin associated proteins was performed as previously described29 using a MOM blocking kit (Vector Labs, Burlingame, CA) and α-sarcoglycan and α-dystroglycan (Novocastra, Newcastle-Upon-Tyne, UK) antibodies (1:100 dilution). Neuronal nitric oxide synthase staining was performed using a goat anti-rabbit antibody (Abcam).
Exon skipping in mdx mouse tissues. Total RNA was extracted from control and treated mouse tissues using TRIzol reagent (Invitrogen) following manufacturer's instructions.
RT-PCR: Four hundred nanograms of RNA template was used in a 50 µl reverse transcription reaction using One Step RT-PCR Kit (Qiagen, Hilden, Germany) and gene-specific primers (Ex 20-26, Fwd: 5′-CAG AAT TCT GCC AAT TGC TGA G-3′, Rev: 5′-TTC TTC AGC TTG TGT CAT CC-3′). Cycle conditions: 50 °C for 30 minutes, followed by 30 cycles of 30 seconds at 94 °C, 1 minute at 58 °C, and 2 minutes at 72 °C. Two microliters of cDNA was further amplified in a 50 µl nested PCR (QIAGEN PCR kit) using the following cycle conditions: 94 °C for 30 seconds, 58 °C for 1 minute, and 72 °C for 1 minute for 24 cycles (Ex 20-26: Fwd: CCC AGT CTA CCA CCC TAT CAG AGC, Rev: CCT GCC TTT AAG GCT TCC TT). PCR products were examined by electrophoresis on a 2% agarose gel.
Quantitative real time PCR: Two micrograms of RNA was reverse transcribed using a High Capacity cDNA Synthesis kit (Applied Biosystems, Branchburg, NJ). Exon skipping qPCR was performed using Syber green Kits (Applied Biosystems), primer sets (IDT) and the StepOne Plus Real-Time PCR system (Applied Biosystems). Primer sets used were as follows: total dystrophin transcripts, ex19-20: Fwd: GCCATAGCACGAGAAAAAGC, Rev: GCATTAACACCCTCATTTGC; Delta23 dmd transcript, Fwd: GCG CTA TCA GGA GAC AAT GAG, Rev: GTT TTT ATG TGA TTC TGT AAT TTC CC.
Plasmids (total dystrophin and delta 23 skipped) were used for the standard curve.
Protein extraction and western blot. Control and treated muscle samples were homogenised in lysis buffer comprising 75 mmol/l Tris–HCl (pH 6.5) and 10% sodium dodecyl sulphate complemented with 5% 2-mercaptoethanol. Samples were heated at 100 °C for 3 minutes before centrifugation and removal of supernatant. Protein levels were measured by Bradford assay (Sigma) and quantified using BSA standards. Ten to fifteen micrograms of protein of untreated and treated mdx sample, and 50% and 10% of these concentrations of C57BL10 protein (positive control) were loaded onto 3–8% Tris-Acetate gels. Proteins were blotted onto polyvinylidene fluoride membrane and probed for dystrophin using DYS1 (Novocastra) and loading control, α-actinin (Sigma), antibodies. Primary antibody was detected by binding of horseradish peroxidase-conjugated anti-mouse immunoglobulin G with lumigen. Western blots were imaged (LiCOR Biosciences, Lincoln, NE) and analyzed using the Odyssey imaging system.
Clinical biochemistry. Plasma samples were taken from the jugular vein of mdx mice immediately following sacrifice by CO2 inhalation. Analysis of toxicity biomarkers was performed by a clinical pathology lab, Mary Lyon Centre, MRC, Harwell, UK.
Statistical analysis. All data reported mean values ± SEM. A multilevel, repeated measures model was implemented for this study. The multilevel statistical approach builds upon traditional statistical methods and is being increasingly implemented in the social, medical and biological sciences.38,39,40,41 The model used for this study takes into account the multiple “relative intensity units” (level 1) for each mouse (level 2) for each treatment (level 3) as performed in the immunohistochemical staining quantification. In this example mdx untreated mice and Pip5e-PMO-treated mice were applied as the constant/fixed parameter, to which the other treatments and wild-type control were compared. This was following a Box-Cox power transformation which was performed to ensure a normal distribution. Statistical analysis was performed using MLwIN version 2.25.
Acknowledgments
This work was supported by research grants to M.J.A.W. from the Muscular Dystrophy Campaign, Duchenne Ireland and the Medical Research Council (G0900887), and joint research grants to M.J.A.W. and M.J.G. from AFM (programme number 14784) and Wellcome Trust/HICF 1009 025. The work in the laboratory of M.J.G. was also supported by the Medical Research Council (MRC programme number U105178803). The authors acknowledge Kay Davies (Department of Physiology, Anatomy and Genetics, University of Oxford, Oxford) for her support and access to animal facilities. The authors thank Daniel Lunn (University of Oxfords Statistics department) for his expert statistical advice, and Tertius Hough (Clinical Pathology Laboratory, Mary Lyon Centre, MRC, Harwell, UK) for his assistance with the clinical biochemistry assays.
Supplementary Material
HPLC chromatogram and MALDI-TOF data for Pip6e-PMO.
qRT-PCR mean values table and quantification of western blots for C57BL10 control, mdx-untreated and Pip6-PMO-treated mdx mice, following a single 12.5 mg/kg, i.v. injection.
Statistical tables for quantitative immunohistochemical staining, qRT-PCR mean values table and quantification of western blots for C57BL10 control, mdx untreated and Pip6e-PMO derivative (Pip6g and Pip6h) treated mdx mice, following a single 12.5 mg/kg, i.v. injection.
Immunohistochemical staining of dystrophin complex proteins in C57BL10 control, mdx untreated, the 5-aa hydrophobic core Pip6-PMO-treated mice.
Toxicity assays assessed in blood samples of C57BL10 control, mdx-untreated, Pip6-PMO- and Pip5e-PMO-treated mdx mice, following a single 12.5 mg/kg, i.v. injection.
REFERENCES
- Emery AE. Population frequencies of inherited neuromuscular diseases–a world survey. Neuromuscul Disord. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- Wilton SD, Lloyd F, Carville K, Fletcher S, Honeyman K, Agrawal S.et al. (1999Specific removal of the nonsense mutation from the mdx dystrophin mRNA using antisense oligonucleotides Neuromuscul Disord 9330–338. [DOI] [PubMed] [Google Scholar]
- Dunckley MG, Manoharan M, Villiet P, Eperon IC., and, Dickson G. Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribonucleotides. Hum Mol Genet. 1998;7:1083–1090. doi: 10.1093/hmg/7.7.1083. [DOI] [PubMed] [Google Scholar]
- Dickson G, Hill V., and, Graham IR. Screening for antisense modulation of dystrophin pre-mRNA splicing. Neuromuscul Disord. 2002;12 Suppl 1:S67–S70. doi: 10.1016/s0960-8966(02)00085-8. [DOI] [PubMed] [Google Scholar]
- Alter J, Lou F, Rabinowitz A, Yin H, Rosenfeld J, Wilton SD.et al. (2006Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology Nat Med 12175–177. [DOI] [PubMed] [Google Scholar]
- Lu QL, Rabinowitz A, Chen YC, Yokota T, Yin H, Alter J.et al. (2005Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles Proc Natl Acad Sci USA 102198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello L, Bassi N, Campagnolo P, Zaccariotto E, Occhi G, Malerba A.et al. (2008In vivo delivery of naked antisense oligos in aged mdx mice: analysis of dystrophin restoration in skeletal and cardiac muscle Neuromuscul Disord 18597–605. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M.et al. (2007Local dystrophin restoration with antisense oligonucleotide PRO051 N Engl J Med 3572677–2686. [DOI] [PubMed] [Google Scholar]
- Goemans NM, Tulinius M, van den Akker JT, Burm BE, Ekhart PF, Heuvelmans N.et al. (2011Systemic administration of PRO051 in Duchenne's muscular dystrophy N Engl J Med 3641513–1522. [DOI] [PubMed] [Google Scholar]
- Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C.et al. (2009Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study Lancet Neurol 8918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K.et al. (2011Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study Lancet 378595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller F, Moseley CF., and, Koreska J. Spinal fusion in Duchenne muscular dystrophy. Dev Med Child Neurol. 1992;34:775–786. doi: 10.1111/j.1469-8749.1992.tb11516.x. [DOI] [PubMed] [Google Scholar]
- English KM., and, Gibbs JL. Cardiac monitoring and treatment for children and adolescents with neuromuscular disorders. Dev Med Child Neurol. 2006;48:231–235. doi: 10.1017/S0012162206000491. [DOI] [PubMed] [Google Scholar]
- Jefferies JL, Eidem BW, Belmont JW, Craigen WJ, Ware SM, Fernbach SD.et al. (2005Genetic predictors and remodeling of dilated cardiomyopathy in muscular dystrophy Circulation 1122799–2804. [DOI] [PubMed] [Google Scholar]
- Wu B, Xiao B, Cloer C, Shaban M, Sali A, Lu P.et al. (2011One-year treatment of morpholino antisense oligomer improves skeletal and cardiac muscle functions in dystrophic mdx mice Mol Ther 19576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malerba A, Sharp PS, Graham IR, Arechavala-Gomeza V, Foster K, Muntoni F.et al. (2011Chronic systemic therapy with low-dose morpholino oligomers ameliorates the pathology and normalizes locomotor behavior in mdx mice Mol Ther 19345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malerba A, Boldrin L., and, Dickson G. Long-term systemic administration of unconjugated morpholino oligomers for therapeutic expression of dystrophin by exon skipping in skeletal muscle: implications for cardiac muscle integrity. Nucleic Acid Ther. 2011;21:293–298. doi: 10.1089/nat.2011.0306. [DOI] [PubMed] [Google Scholar]
- Townsend D, Yasuda S, Li S, Chamberlain JS., and, Metzger JM. Emergent dilated cardiomyopathy caused by targeted repair of dystrophic skeletal muscle. Mol Ther. 2008;16:832–835. doi: 10.1038/mt.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas JM, Nye DJ, Henley JB., and, Plochocki JH. Voluntary exercise induces structural remodeling in the hearts of dystrophin-deficient mice. Muscle Nerve. 2010;42:881–885. doi: 10.1002/mus.21783. [DOI] [PubMed] [Google Scholar]
- Mitchell DJ, Kim DT, Steinman L, Fathman CG., and, Rothbard JB. Polyarginine enters cells more efficiently than other polycationic homopolymers. J Pept Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- Abes R, Arzumanov A, Moulton H, Abes S, Ivanova G, Gait MJ.et al. (2008Arginine-rich cell penetrating peptides: design, structure-activity, and applications to alter pre-mRNA splicing by steric-block oligonucleotides J Pept Sci 14455–460. [DOI] [PubMed] [Google Scholar]
- Abes R, Moulton HM, Clair P, Yang ST, Abes S, Melikov K.et al. (2008Delivery of steric block morpholino oligomers by (R-X-R)4 peptides: structure-activity studies Nucleic Acids Res 366343–6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulton HM, Fletcher S, Neuman BW, McClorey G, Stein DA, Abes S.et al. (2007Cell-penetrating peptide-morpholino conjugates alter pre-mRNA splicing of DMD (Duchenne muscular dystrophy) and inhibit murine coronavirus replication in vivo Biochem Soc Trans 35Pt 4826–828. [DOI] [PubMed] [Google Scholar]
- Jearawiriyapaisarn N, Moulton HM, Sazani P, Kole R., and, Willis MS. Long-term improvement in mdx cardiomyopathy after therapy with peptide-conjugated morpholino oligomers. Cardiovasc Res. 2010;85:444–453. doi: 10.1093/cvr/cvp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Moulton HM, Iversen PL, Jiang J, Li J, Li J.et al. (2008Effective rescue of dystrophin improves cardiac function in dystrophin-deficient mice by a modified morpholino oligomer Proc Natl Acad Sci USA 10514814–14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F, Joliot A, Bloch-Gallego E, Zahraoui A, Triller A., and, Prochiantz A. Antennapedia homeobox as a signal for the cellular internalization and nuclear addressing of a small exogenous peptide. J Cell Sci. 1992;102 (Pt 4):717–722. doi: 10.1242/jcs.102.4.717. [DOI] [PubMed] [Google Scholar]
- Turner JJ, Ivanova GD, Verbeure B, Williams D, Arzumanov AA, Abes S.et al. (2005Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans-activation in cells Nucleic Acids Res 336837–6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova GD, Arzumanov A, Abes R, Yin H, Wood MJ, Lebleu B.et al. (2008Improved cell-penetrating peptide-PNA conjugates for splicing redirection in HeLa cells and exon skipping in mdx mouse muscle Nucleic Acids Res 366418–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Saleh AF, Betts C, Camelliti P, Seow Y, Ashraf S.et al. (2011Pip5 transduction peptides direct high efficiency oligonucleotide-mediated dystrophin exon skipping in heart and phenotypic correction in mdx mice Mol Ther 191295–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Moulton HM, Seow Y, Boyd C, Boutilier J, Iverson P.et al. (2008Cell-penetrating peptide-conjugated antisense oligonucleotides restore systemic muscle and cardiac dystrophin expression and function Hum Mol Genet 173909–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Moulton HM, Betts C, Seow Y, Boutilier J, Iverson PL.et al. (2009A fusion peptide directs enhanced systemic dystrophin exon skipping and functional restoration in dystrophin-deficient mdx mice Hum Mol Genet 184405–4414. [DOI] [PubMed] [Google Scholar]
- Saleh AF, Arzumanov AA, Yin H, Betts C, Wood MJA., and, Gait MJ.et al. Enhancement of exon skipping and dystrophin production by 3'-peptide conjugates of morpholino (PMO) oligonucleotides in a mdx mouse model of Duchenne muscular dystrophy in Collection Symposium Series, Chemistry of Nucleic Acid Components. Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic: Prague, 2011, pp. 292–296 [Google Scholar]
- Saleh AF, Arzumanov A, Abes R, Owen D, Lebleu B., and, Gait MJ. Synthesis and splice-redirecting activity of branched, arginine-rich peptide dendrimer conjugates of peptide nucleic acid oligonucleotides. Bioconjug Chem. 2010;21:1902–1911. doi: 10.1021/bc100275r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arechavala-Gomeza V, Kinali M, Feng L, Brown SC, Sewry C, Morgan JE.et al. (2010Immunohistological intensity measurements as a tool to assess sarcolemma-associated protein expression Neuropathol Appl Neurobiol 36265–274. [DOI] [PubMed] [Google Scholar]
- Malerba A, Thorogood FC, Dickson G., and, Graham IR. Dosing regimen has a significant impact on the efficiency of morpholino oligomer-induced exon skipping in mdx mice. Hum Gene Ther. 2009;20:955–965. doi: 10.1089/hum.2008.157. [DOI] [PubMed] [Google Scholar]
- Gupta A, Mandal D, Ahmadibeni Y, Parang K., and, Bothun G. Hydrophobicity drives the cellular uptake of short cationic peptide ligands. Eur Biophys J. 2011;40:727–736. doi: 10.1007/s00249-011-0685-4. [DOI] [PubMed] [Google Scholar]
- Thomas CR, K.E.M.B., Graham M, Samir ELA, Caroline G, Corinne B.et al. (2012Wood, Expression analysis in multiple muscle groups and serum reveals complexity in the microRNA transcriptome of the mdx mouse with implications for therapy Nucleic Acid Ther [DOI] [PMC free article] [PubMed]
- Butterfield A, Vedagiri V, Lang E, Lawrence C, Wakefield MJ, Isaev A.et al. (2004PyEvolve: a toolkit for statistical modelling of molecular evolution BMC Bioinformatics 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks G, Dolphin M, Vanbeveren P., and, Hart DL. Referral source and outcomes of physical therapy care in patients with low back pain. J Orthop Sports Phys Ther. 2012. [DOI] [PubMed]
- Bernier J, Feng Y., and, Asakawa K. Strategies for handling normality assumptions in multi-level modeling: a case study estimating trajectories of Health Utilities Index Mark 3 scores. Health Rep. 2011;22:45–51. [PubMed] [Google Scholar]
- Winter EM, Eston RG, Lamb KL. Statistical analyses in the physiology of exercise and kinanthropometry. J Sports Sci. 2001;19:761–775. doi: 10.1080/026404101317015429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HPLC chromatogram and MALDI-TOF data for Pip6e-PMO.
qRT-PCR mean values table and quantification of western blots for C57BL10 control, mdx-untreated and Pip6-PMO-treated mdx mice, following a single 12.5 mg/kg, i.v. injection.
Statistical tables for quantitative immunohistochemical staining, qRT-PCR mean values table and quantification of western blots for C57BL10 control, mdx untreated and Pip6e-PMO derivative (Pip6g and Pip6h) treated mdx mice, following a single 12.5 mg/kg, i.v. injection.
Immunohistochemical staining of dystrophin complex proteins in C57BL10 control, mdx untreated, the 5-aa hydrophobic core Pip6-PMO-treated mice.
Toxicity assays assessed in blood samples of C57BL10 control, mdx-untreated, Pip6-PMO- and Pip5e-PMO-treated mdx mice, following a single 12.5 mg/kg, i.v. injection.