Figure 1.
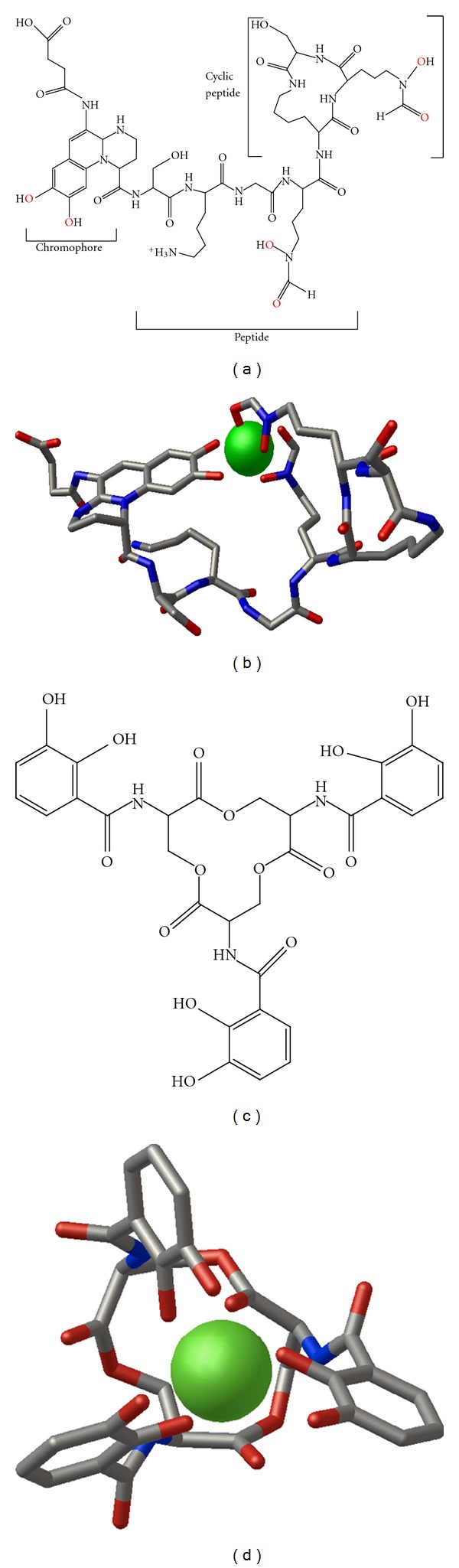
Molecular structures of ferric bacterial siderophores used in this study. (a) Chemical structure of a group I pyoverdine from bacterial species Pseudomonas fluorescens (strain ATCC 13525) illustrating its derivatized peptide structure. The pyoverdine amino acid sequence is Ser*-Lys-Gly-FHO-cyclic [Lys-FHO-Ser*], where Ser* is D-serine and FHO is N-formyl-N-hydroxy-ornithine. Oxygens in red represent atoms that coordinate iron in the 3D structures. (b) Ferric pyoverdine structure taken from the 3D structure of a pyoverdine-Fe transporter protein (FpvA) in complex with pyoverdine (ATCC 13525) and Fe (PDB 2W78). Only the pyoverdine and iron (green) atoms are shown. Both hydroxyl groups on the chromophore and FHO side chain oxygens form interactions with iron. (c) Chemical structure of enterobactin. (d) Ferric enterobactin structure isolated from the 3D structure of a mutant of NGAL in complex with ferric enterobactin (PDB 3CMP). Illustrations for (b) and (d) were made using Python Molecular Viewer 1.5.4. Each ligand is shown as a stick figure with coloring by atom type. Iron atoms are shown in green space-filling representation.
