Figure 5.
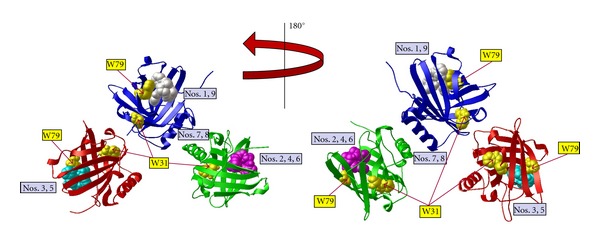
Enterobactin binds to NGAL. The docking of ferric enterobactin into the NGAL crystal structure (PDB 1L6M) using AutoDock Vina was performed as a positive control to validate the pyoverdine docking simulation. NGAL crystal structure with each monomer presented in ribbon diagram colored red, blue, and green, plus ferric enterobactin. AutoDock Vina found nine positions in which ferric enterobactin docked, seven of which were in one of the calyx of one of the monomers, that is, the ligand-binding site. The two remaining positions, modes 7 and 8, are between the blue and green monomers (enterobactin is not shown in modes 7 and 8, for clarity). For this illustration only one ferric enterobactin molecule is displayed in each of the predicted binding positions on the rigid NGAL. Tryptophan residues W31 and W79 are highlighted in yellow. The docking of ferric enterobactin to NGAL view was rotated 180° for display in the view at right. The binding affinity for each docking mode is listed in Table 1.
