Abstract
PEGylation is the covalent attachment of polyethylene glycol to proteins, and it can be used to alter immunogenicity, circulating half life and other properties of therapeutic proteins. To determine the impact of PEGylation on protein conformation, we applied hydrogen/deuterium exchange mass spectrometry (HDX MS) to analyze Granulocyte Colony Stimulating Factor (G-CSF) upon PEGylation as a model system. The combined use of HDX automation technology and data analysis software allowed reproducible and robust measurements of the deuterium incorporation levels for peptic peptides of both PEGylated and non-PEGylated G-CSF. The results indicated that significant differences in deuterium incorporation were induced by PEGylation of G-CSF, although the overall changes observed were quite small. PEGylation did not result in gross conformational rearrangement of G-CSF. The data complexity often encountered in HDX MS measurements was greatly reduced though a data processing and presentation format designed to facilitate the comparison process. This study demonstrates the practical utility of HDX MS for comparability studies, process monitoring and protein therapeutic characterization in the biopharmaceutical industry.
Keywords: Protein conformation, polyethylene glycol, comparability, biopharmaceutical, post-translational modification, therapeutic protein
Introduction
Since Davis and Abuchowski discovered the methodology of covalent attachment of PEG (polyethylene glycol) to therapeutic proteins in the late 1970s [1], PEGylation has become an effective strategy to improve the pharmacokinetic behavior of proteins. Some of the benefits PEGylation affords include reduced immunogenicity, antigenicity, and prolonged circulating time [2]. A number of protein drugs are improved by PEGylation, among which many have been approved by the FDA and already reached the market [3, 4], and many more have made it to late phase clinical trials. Covalent attachment of PEG may cause possible conformational changes, steric interferences, and changes in electrostatic-binding properties for certain proteins [5]. Binding affinities to target proteins may also be affected by these physicochemical changes resulting in reduced in vitro activity, as reviewed elsewhere [6]. Thus it is of great importance to effectively and stringently characterize the changes that PEG may cause in a protein, including changes in protein conformation. This information is needed to ensure activity and manufacturing reproducibility for regulatory approval.
It is, however, quite analytically challenging to characterize conformational changes of such molecules, due to a number of factors, including: the complexity of protein structure (with several levels of order), the high degree of heterogeneity of PEG, the number of PEG moieties that can potentially be attached, and the site(s) of PEGylation. In addition, product specification and in-process testing are complicated by the presence of the PEG polymer, hindering process development and stability studies if the analytical method is not up to the task. To date there are a limited number of examples in the scientific literature describing approaches for the structural characterization of PEGylated biomolecules. Techniques such as gel electrophoresis [7], chromatography [8] and mass spectrometry [9, 10] have been employed but were limited to the characterization of the primary structures of PEGylated biomolecules. X-ray crystallography and nuclear magnetic resonance (NMR) determine protein structure with high spatial resolution, however, the technologies require either lengthy crystallization processes or isotopically labeled proteins, and are therefore inhibitory for routine sample analysis. Crystallography is limited by the crystallization properties of the molecule and NMR by the size of the proteins analyzed, both of which are especially challenging for PEGylated proteins.
Hydrogen/deuterium exchange mass spectrometry (HDX MS) is an orthogonal biophysical technique which has seen profound advances for the study of protein conformation and dynamics [11, 12]. The technique requires only small quantity of sample (picomoles) for solution phase studies and can provide useful results with dilute solutions (sub-micromolar). It can be used to study proteins that are hard to purify and can reveal protein conformational dynamics on a wide time scale. With the aid of pepsin digestion, which cleaves the protein into small peptides after the labeling is quenched, deuterium incorporation can be localized to short (5–10 amino acids) stretches of the primary structure [13]. The technique has been successfully applied to the study of protein structure dynamics for large molecules such as antibodies [14], the characterization of protein-ligand interactions [15, 16] and epitope mapping of protein-protein interactions [17, 18]. Given the potential for HDX MS to provide conformational information for proteins not amenable to classical structural tools, we sought to determine the utility of HDX MS for analysis of PEGylated proteins.
In this paper we demonstrate how we utilized a standardized LC-MS workflow dedicated to HDX at the peptide level to efficiently and effectively compare the conformational dynamics of a PEGylated biopharmaceutical drug [granulocyte colony stimulating factor (G-CSF)] to the conformational dynamics of the non-PEGylated form. G-CSF has become an important cytokine for medical treatment of patients suffering from granulopoenia [19]. By conjugating a 20 kDa PEG to the N-terminal alpha amino group, fast blood clearance of G-CSF was reduced and there was an improved pharmacokinetic profile [20]. Our results identify local conformational differences within G-CSF that are induced by PEGylation. Finally, we demonstrate the use of a reduced comparability format using a new software tool designed for comparability measurements of proteins.
Experimental
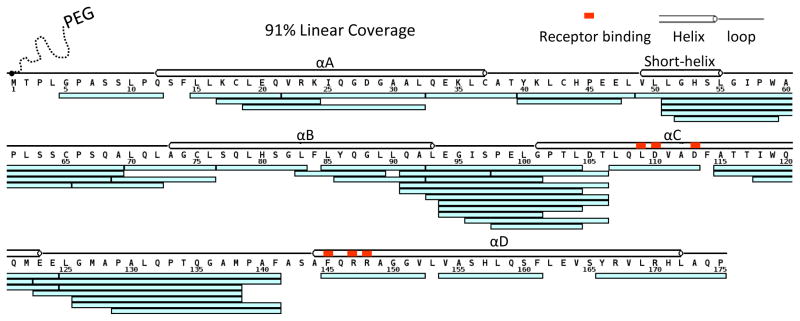
PEGylated G-CSF was manufactured by GenScript Inc (Piscataway, NJ, USA) by attaching a 20 kDa methoxypolyethylene glycol propionaldehyde (mPEG-ALD) to the N-terminal amino acid of G-CSF. Each protein was reconstituted to 25.7 μM in 50 mM potassium phosphate in 100% H2O, pH 7.00, then diluted 20-fold with either 50 mM potassium phosphate in 100% H2O, pH 7.00 for the undeuterated experiments, or 50 mM potassium phosphate in 99.9% D2O, pD 7.00 for the deuterated experiments. Note that measured pH values of D2O solutions (also known as pH*) were all adjusted to the corresponding pD values through use of the well-known equation (pD = pHread + 0.40) [21, 22]. After dilution, the samples were incubated at room temperature for various amounts of time (0 sec for undeuterated experiments, and 10 sec, 1, 12, 60 and 240 min for deuterated experiments), then quenched by reducing the pH to 2.50 with a 1:1 dilution with ice-cold quench buffer [200 mM potassium phosphate, 1.5 M guanidine HCl (Pierce, Thermo Scientific, Rockford, IL USA), 500 mM (tris(2-carboxyethyl)phosphine) (Sigma, C4706) in H2O, pH 2.40]. Quenched samples (32 pmoles for each time point) were immediately injected into a nanoACQUITY UPLC™ system with HDX Technology (Waters Corporation, Milford, MA, USA) [23]. The HDX manager of this system provides temperature control for UPLC separation at 0 °C. The online digestion was performed using an immobilized pepsin column, 2.0 × 30 mm (Applied Biosystems, CA, USA) for 4 min in 0.05% formic acid, H2O at a flow rate of 75 μL/min. The entire digestion was held at 20 °C inside of the temperature controlled digestion column compartment of the HDX manager. Peptides were trapped and desalted online using an ACQUITY UPLC® BEH C18 1.7 μm VanGuard™ Pre-column (Waters) at 0 °C. The flow was diverted by switching valves and trapped peptides were eluted into an ACQUITY UPLC BEH C18 1.7 μm, 1 mm × 100 mm column (Waters) held at 0 °C. Peptides were separated with a 7 min linear acetonitrile gradient (7–40%) containing 0.1% formic acid at a flow rate of 40 μL/min. The eluent was directed into a SYNAPT® G2 HDMS mass spectrometer (Waters) with electrospray ionization and lock-mass correction (using Glu-fibrinogen peptide). Mass spectra were acquired in MSE mode over the m/z range of 50–2000. Blank injections were performed between each sample injection to confirm the absence of peptide carryover from previous runs [24]. Peptides of both G-CSF and PEGylated G-CSF were identified in the undeuterated sample using ProteinLynx Global Server software (Waters). The sequence coverage map of G-CSF (Figure 1) was plotted using the online tool MSTools [25]. The deuterium incorporation levels for each peptic peptide were automatically calculated using DynamX software (Waters) with the algorithm described elsewhere [26].
Fig. 1.
Peptic peptides of G-CSF and PEGylated G-CSF. Each cyan bar under the sequence indicates a common peptide reproducibly identified in both proteins. Secondary structural information and receptor binding sites are also illustrated in the map.
Three undeuterated experiments and two complete HDX experiments were performed for each protein (10 separate experiments in total for all forms of G-CSF) within two consecutive days. Two additional complete HDX experiments were repeated for PEGylated G-CSF to assess the uncertainty of the measurements. As these were relative experiments in which we compared deuterium incorporation into two states of the same proteins, no 100% deuterated reference was prepared and no adjustment was made for deuterium back-exchange during deuterium uptake calculation; therefore all results are reported as relative deuterium level as described elsewhere [11]. The data are expressed in either mass units (Da) or relative fractional exchange, which was calculated by dividing the deuterium level (in Da) by the total number of backbone amide hydrogens that could have become deuterated (equal to the number of amino acids, minus proline residues minus 1 for the N-terminal amide); see also Ref. [27] for more details). Based on data obtained from the two HDX experiments and the additional two replicate experiments over different days (n = 4), the experimental uncertainty of measuring a deuterium level was found to be ±0.15 Da which is similar to the uncertainty reported elsewhere [27, 28]. It has been previously reported that this uncertainly appears to be independent of the magnitude of peptide size, HDX labeling time or the magnitude of the mass difference [27]. Using this estimate of experimental uncertainty, and a 98% confidence interval, a significant difference value of 0.5 Da was calculated. If the difference in HDX level for any peptide at any labeling time exceeded 0.5 Da (either positive difference or negative difference), then it was considered a significant difference. Similarly, for each peptide, the summed value of HDX differences across all labeling time points (10 sec, 1, 12, 60 and 240 min) was used to calculate (again, with a 98% confidence interval) a value of 1.5 Da as a significant difference value for summed HDX differences. Interpretation of a significant difference was similar to that for the unsummed difference above: if the summed differences for any peptide exceeded 1.5 Da (either positively or negatively), it was considered a significant difference. The significance differences are indicated on the butterfly and difference charts created with the plotting algorithms described in detail by Houde et al. [27]. For a comparison analysis of HDX results between G-CSF and PEGylated C-CSF, the butterfly chart and the difference chart were made directly by the DynamX software.
Results and Discussion
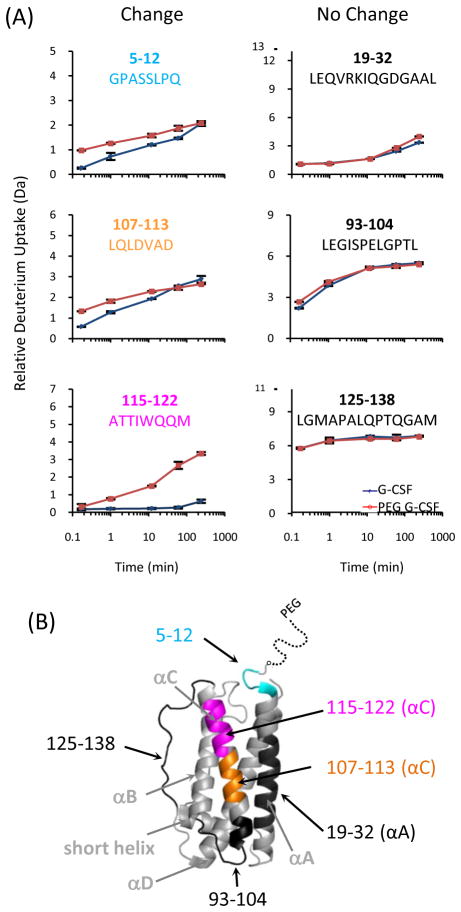
We sought to determine if differences in uptake would be found at or near the receptor binding sites [29] (orange in Figure 1), thereby indicating if PEGylation led to altered biological activity of the protein. In order to compare HDX between G-CSF and PEGylated G-CSF, it was important to monitor common peptic peptides that were produced during digestion of either protein. Unveiling different deuterium uptake in peptic peptides produced after the labeling reaction had been quenched provided location-specific information about protein conformation and dynamics. Well controlled, and preferably highly automated, digestion conditions generate the same peptic peptides in a very reproducible fashion. A total of six online digestion experiments (three for each protein) were performed for undeuterated G-CSF and PEGylated G-CSF as outlined in the Experimental section. The chromatographic separations were highly reproducible and the mass spectra had a high signal-to-noise ratio for each peptide identified (see Supplemental Figure S1). Forty-six peptic peptides were unambiguously identified in each protein in at least two of the three runs, constituting a linear sequence coverage of 91% of the peptide backbone of G-CSF, with overlapping peptides in multiple regions (Figure 1). The high number of overlapping peptides allowed us to determine deuterium incorporation in shorter regions, by virtue of difference analysis between overlapping peptides. After the peptic peptides were identified, both G-CSF and PEGylated G-CSF were subjected to HDX MS analysis and the deuterium levels of the forty-six peptides that were reproducibly observed in the undeuterated samples were measured in duplicate for each labeling time-point and form of G-CSF (see Supplemental Figure S2). Each plot represents the average relative deuterium uptake of each peptide at each time point, determined by averaging the deuterium level found in each separate replicate experiment. There were some peptides that displayed differences in deuterium incorporation between the two forms of G-CSF, and some that showed no differences. Figure 2A illustrates representative data of peptides in which deuterium uptake was the same or different. The location of each of the peptides was overlaid (Figure 2B) onto the three-dimensional X-ray crystal structure of G-CSF (PDB code 2D9Q) [29].
Fig. 2.
Deuterium uptake curves of six peptic peptides (A), three that had changes upon PEGylation (left) and three that did not (right). The peptides are illustrated on the G-CSF crystal structure (B), with the color of the peptide matching the color of the residue labels in part A: cyan, residues 5-12; orange, residues 107-113; pink, residues 115-122; black residues 19-32, 93-104 and 125-138. The three-dimensional structure of G-CSF is from PDB code 2D9Q [29].
The results suggested that multiple locations in the protein structure had changes in deuterium level (> 0.5 Da) between the two forms of the protein. For example, small but significant change to exchange levels (~1.0 Da difference at10 sec of labeling) was seen in peptide GPASSLPQ (residues 5-12) in the PEGylated form compared to the non-PEGylated form. The beginning of this peptide is located four residues away from the PEGylation site (Met1). Other examples of increased deuterium incorporation in the PEGylated form include the peptides LFLYQGL (residues 83-89) and TTIWQQM (residues 115-122). These peptides are located in alpha-helix regions, where the non-PEGylated form showed minimum uptake throughout the labeling time-course [the neighboring overlapping peptide 115-124 (see Figure S2) showed similar deuterium incorporation]. Interestingly, peptide 115-122 is located close to the PEGylation site in the crystal structure (see Figure 2B). A subtle increase in deuteration was also seen for peptide LQLDVAD (residues 107-113) in the PEGylated form. Comparing peptide 115-122 and 107-113, which are both part of the same long alpha-helix, the C-terminal portion of the helix (residues 115-122) close to the PEG site showed relatively more increase in deuteration than the N-terminal portion (residues 107-113). For parts of G-CSF located in alpha-helices, their backbone amide hydrogens are involved in the formation of hydrogen bonds which should be quite protected from HDX. The increased amount of deuteration in the PEGylated form suggests that these amide hydrogens are more prone to exchange upon PEGylation, especially in regions closer to the site of PEGylation. These observations may also reflect differences in conformational stability between PEGylated and non-PEGylated G-CSF.
Although small but significant differences in deuteration were found in multiple regions of G-CSF, for 65% of G-CSF, there were no detectable differences in deuterium incorporation upon PEGylation. Many such peptides were located in the loop regions (e.g., residues 125-138, as shown in Figure 2 and its overlapped peptides), where exchange is predicted to be rapid. The peptide covering residues 19-32 (part of an alpha-helix) indicated no difference to deuteration and remained low in deuterium uptake for both forms. There are multiple amino acids in this peptide that are involved in receptor binding (see Figure 1). Other peptides containing amino acids known to be involved in receptor binding (e.g. residues 107-113, residues 145-152) showed only minor differences in deuterium incorporation levels across all the labeling times. Therefore, by HDX measurements, the conformation and conformational stability of the receptor binding regions of the protein were not significantly affected by PEGylation. This result is consistent with G-CSF maintaining most of its biological activity after PEGlyation, as has been reported elsewhere [30].
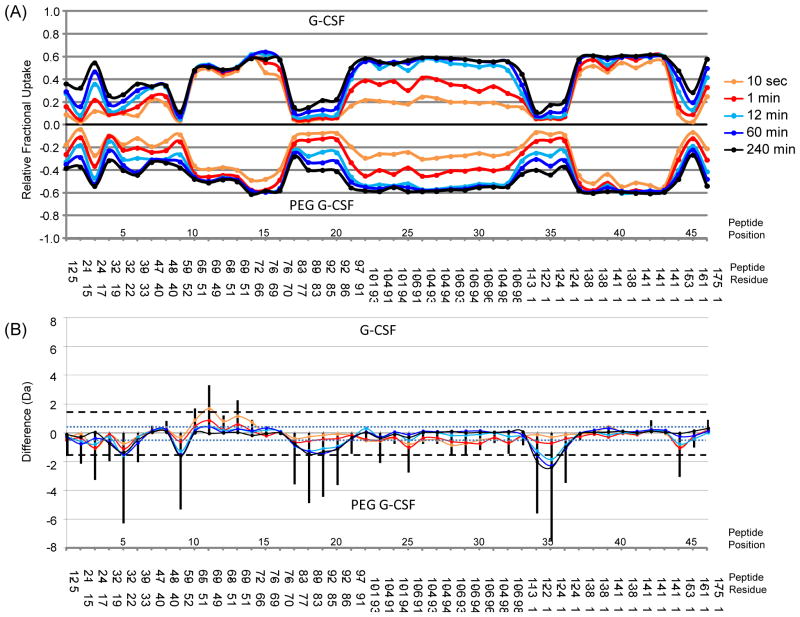
While there are multiple ways to display the complex HDX profiles obtained in a comparison experiment (for example, see Supplementary Figure S3), we advocate visualization of such HDX data in a comparability butterfly chart (Figure 3A) and a difference chart (Figure 3B). This form of data presentation allows rapid qualitative and quantitative analysis of differences and the location of such differences. This plotting scheme was recently introduced by Houde et al. [27] and was used to compared a bound vs. a free form of N-myristoyltransferase [31]. Both the butterfly and difference charts compare the deuterium incorporation levels of all peptides between G-CSF and PEGylated G-CSF. The data in the butterfly chart represent the average relative fractional exchange (here, the average of two independent experimental determinations) for all peptides in each labeling time-point. From this chart, one can easily compare the relative deuterium incorporation across the entire polypeptide backbone with both spatial and temporal information. The chart shows that some regions of G-CSF were much more rapidly deuterated than others and changes in the rate of deuteration are also evident. For example, peptides between residues 123 and 141 (peptide position 37 to 43) were deuterated quickly resulting in greater than 50% of amide hydrogens already exchanging after only 10 sec of deuterium labeling. Multiple overlapping peptides found between residues 83 and 97 (peptide position at 18 to 21) show differences in the rate of deuteration between the PEGylated and non-PEGylated forms of G-CSF, with the PEGylated form becoming more deuterated sooner.
Fig. 3.
Comparability between PEGylated and non-PEGylated G-CSF. A butterfly chart of the raw deuterium levels (A) and a difference chart (B) compare the forty-six common peptides (along the x-axis). The color code for the time points is shown at the top right. Relative fractional uptake was calculated by dividing the deuterium level (in Da) by the total number of backbone amide hydrogens that could have become deuterated (equal to the number of amino acids, minus proline residues minus 1 for the N-terminal amide) See also Ref. [27] for more details. In (B), the blue dotted line is set at 0.5 Da (both positive and negative difference) indicating the threshold for significant differences in raw fractional uptake (colored lines, code same as in part A). The black vertical bars indicate the summed differences in deuteration for each peptide and the black dotted line is set at 1.5 Da (positive and negative) to indicate the threshold for a significant difference in summed deviations. See also Ref. [27] for a more in-depth explanation of these graphs and the algorithms used to create them.
For comparison purposes, a difference chart (Figure 3B) was created to highlight where changes occurred between the two forms of G-CSF. A negative value in this chart, for example at peptide position 35 (residues 115-124), indicates that this region had become more deuterated in PEGylated G-CSF compared to the same peptide from non-PEGylated G-CSF. There are two parts to the difference graph, plotted together. The first is the raw difference (the colored lines) which represents the difference in HDX for each peptide at each time-point. The raw difference in peptide 35 is considered to be significant because it exceeded the threshold value of 0.5 Da (blue dotted line). The second part of the difference graph is the solid vertical bar which represents the summed differences of all time-points in the labeling time course for a corresponding peptide. When the value of the vertical bar exceeds 1.5 Da, it is considered to be a significant difference.
It is clear that there are multiple regions along G-CSF that showed significant differences in deuterium incorporation between the two forms. Both the raw difference values (colored lines) and the summed differences for each peptide (the vertical black bars) exceed significance lines for many peptides. In most cases the difference values are negative, indicating that in this particular graph with this particular plotting (G-CSF on top, PEG G-CSF on bottom) the deuteration is greater for these peptides in PEGylated G-CSF than the non-PEGylated form. A few regions showed positive difference values (e.g., peptide 11-13 corresponding to amino acid residues 49-69) meaning deuteration was less in the PEGylated form. As controls, difference graphs were also created for totally separate, duplicate runs of G-CSF (or PEGylated G-CSF), to assess experimental variability (Supplementary Figure S4). Very minimal differences were observed in these plots, illustrating the good reproducibility and consistency of the analyses, and demonstrating that the differences observed in the plots of PEGylated vs. non-PEGylated reflected measurable differences in solution behavior.
Conclusions
These results indicate that small changes in protein conformation were induced by PEGylation of G-CSF and that these changes were detectable by HDX MS. The changes were localized to various parts of the protein. Although most of the differences observed were significant from a statistical and analytical standpoint, overall, the changes observed were quite small. Changes of a few Daltons in several peptides does not equate with massive protein conformational changes, unfolding or structural rearrangements. Therefore, for G-CSF, PEGylation does not drastically alter the conformation of the protein. This is consistent with other findings in which PEGylation does not lead to conformational changes of the polypeptide chain [27, 32]. Previous studies of G-CSF in non-PEGylated and PEGylated form have noted presumed changes to the oligomerization status of G-CSF depending on PEGylation [30, 33]. In one of those studies[33], the concentrations investigated (5 mg/mL) were not the same as in our work (<1 mg/mL) and differences in oligomerization were observed after prolonged exposure at 37 °C while the proteins were monomers in solution prior to elevated temperature. By contrast, under the conditions in our experiments, there was no exposure to elevated temperature and the proteins were at much lower concentration. Our results show that the peptide containing residues 115-122 had more deuterium incorporation for the PEGlyated form, which was significantly different from the control. If the non PEGylated form existed as a dimer that was disrupted by the presence of PEG, one would see increased deuteration of the PEGylated form – which is exactly what we observed. Our results are therefore consistent with hypotheses involving changes to the oligomeric status of G-CSF is altered by PEGylation.
For other proteins besides G-CSF, there have been studies which demonstrate that PEGylation had some impact on protein conformation and even activity. For example, PEGylation of Val-1 in the α chain of hemoglobin destabilizes the tetrametric structure [34], PEGylation of cholesterol oxidase changed its substrate specificity from total cholesterol to high density lipoprotein cholesterol, and PEGylation of a modified version of human growth hormone (pegvisomant) changed the activity from agonist to antagonist [35].
We conclude that the conformational differences between the PEGylated and non-PEGlyated forms of G-CSF observed in our study are real, but may not have significant impact on the biological activity, especially given that differences in HDX levels observed in the epitope regions of the protein were essentially nonexistent. Finally, this study illustrates that if there were changes in conformation in a protein as the result of some modification, process change, or other outside forces, they could be revealed by HDX MS. HDX MS performed at the peptide level is a powerful tool that not only revealed detailed changes in protein structure induced by PEGylation, but also localized and quantified the extent of the changes. The experiments and data analysis were completed within a couple of days with automation in labeling, data collection and data processing. Data complexity was also greatly reduced through a data presentation format designed to facilitate comparison studies. With such capabilities, HDX MS is ideally suited to assess protein conformation and dynamics. As illustrated in this study, it has great potential in the biopharmaceutical industry for comparability studies, process monitoring and protein therapeutic characterization.
Supplementary Material
Acknowledgments
Authors would like to thank Dr. Damian Houde (Biogen-Idec) for helpful discussions. This work was supported in part by a grant from the NIH (GM086507). This is contribution 994 from the Barnett Institute.
References
- 1.Abuchowski A, McCoy JR, Palczuk NC, van Es T, Davis FF. Effect of covalent attachment of polyethylene glycol on immunogenicity and circulating life of bovine liver catalase. J Biol Chem. 1977;252:3582–3586. [PubMed] [Google Scholar]
- 2.Greenwald RB, Choe YH, McGuire J, Conover CD. Effective drug delivery by PEGylated drug conjugates. Adv Drug Deliv Rev. 2003;55:217–250. doi: 10.1016/s0169-409x(02)00180-1. [DOI] [PubMed] [Google Scholar]
- 3.Pasut G, Veronese FM. PEG conjugates in clinical development or use as anticancer agents: an overview. Adv Drug Deliv Rev. 2009;61:1177–1188. doi: 10.1016/j.addr.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Veronese FM, Mero A. The impact of PEGylation on biological therapies. BioDrugs. 2008;22:315–329. doi: 10.2165/00063030-200822050-00004. [DOI] [PubMed] [Google Scholar]
- 5.Reddy KR. Controlled-release, pegylation, liposomal formulations: new mechanisms in the delivery of injectable drugs. Ann Pharmacother. 2000;34:915–923. doi: 10.1345/aph.10054. [DOI] [PubMed] [Google Scholar]
- 6.Chapman AP. PEGylated antibodies and antibody fragments for improved therapy: a review. Adv Drug Deliv Rev. 2002;54:531–545. doi: 10.1016/s0169-409x(02)00026-1. [DOI] [PubMed] [Google Scholar]
- 7.Na DH, Park EJ, Youn YS, Moon BW, Jo YW, Lee SH, Kim WB, Sohn Y, Lee KC. Sodium dodecyl sulfate-capillary gel electrophoresis of polyethylene glycolylated interferon alpha. Electrophoresis. 2004;25:476–479. doi: 10.1002/elps.200305684. [DOI] [PubMed] [Google Scholar]
- 8.Wang YS, Youngster S, Grace M, Bausch J, Bordens R, Wyss DF. Structural and biological characterization of pegylated recombinant interferon alpha-2b and its therapeutic implications. Adv Drug Deliv Rev. 2002;54:547–570. doi: 10.1016/s0169-409x(02)00027-3. [DOI] [PubMed] [Google Scholar]
- 9.Huang L, Gough PC, Defelippis MR. Characterization of poly(ethylene glycol) and PEGylated products by LC/MS with postcolumn addition of amines. Anal Chem. 2009;81:567–577. doi: 10.1021/ac801711u. [DOI] [PubMed] [Google Scholar]
- 10.Cindric M, Cepo T, Galic N, Bukvic-Krajacic M, Tomczyk N, Vissers JP, Bindila L, Peter-Katalinic J. Structural characterization of PEGylated rHuG-CSF and location of PEG attachment sites. J Pharm Biomed Anal. 2007;44:388–395. doi: 10.1016/j.jpba.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 11.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 12.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houde D, Arndt J, Domeier W, Berkowitz S, Engen JR. Characterization of IgG1 Conformation and Conformational Dynamics by Hydrogen/Deuterium Exchange Mass Spectrometry. Anal Chem. 2009;81:5966. doi: 10.1021/ac9009287. [DOI] [PubMed] [Google Scholar]
- 15.Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 16.Zhu MM, Rempel DL, Zhao J, Giblin DE, Gross ML. Probing Ca2+-induced conformational changes in porcine calmodulin by H/D exchange and ESI-MS: effect of cations and ionic strength. Biochem. 2003;42:15388–15397. doi: 10.1021/bi035188o. [DOI] [PubMed] [Google Scholar]
- 17.Baerga-Ortiz A, Hughes CA, Mandell JG, Komives EA. Epitope mapping of a monoclonal antibody against human thrombin by H/D-exchange mass spectrometry reveals selection of a diverse sequence in a highly conserved protein. Protein Sci. 2002;11:1300–1308. doi: 10.1110/ps.4670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coales SJ, Tuske SJ, Tomasso JC, Hamuro Y. Epitope mapping by amide hydrogen/deuterium exchange coupled with immobilization of antibody, on-line proteolysis, liquid chromatography and mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:639–647. doi: 10.1002/rcm.3921. [DOI] [PubMed] [Google Scholar]
- 19.Nagata S, Tsuchiya M, Asano S, Kaziro Y, Yamazaki T, Yamamoto O, Hirata Y, Kubota N, Oheda M, Nomura H, et al. Molecular cloning and expression of cDNA for human granulocyte colony-stimulating factor. Nature. 1986;319:415–418. doi: 10.1038/319415a0. [DOI] [PubMed] [Google Scholar]
- 20.Kinstler O, Molineux G, Treuheit M, Ladd D, Gegg C. Mono-N-terminal poly(ethylene glycol)-protein conjugates. Adv Drug Deliv Rev. 2002;54:477–485. doi: 10.1016/s0169-409x(02)00023-6. [DOI] [PubMed] [Google Scholar]
- 21.Glasoe PK. Use of glass electrodes to measure acidities in deuterium oxide. J Phys Chem. 1960;64:188–189. [Google Scholar]
- 22.Connelly GP, Bai Y, Jeng MF, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 23.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang J, Rand KD, Beuning PJ, Engen JR. False EX1 signatures caused by sample carryover during HX MS analyses. Int J Mass Spectrom. 2011;302:19–25. doi: 10.1016/j.ijms.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavana D, Mana P. MSTools—Web based application for visualization and presentation of HXMS data. Int J Mass Spectrom. 2011;302:53–58. [Google Scholar]
- 26.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 27.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2011;100:2071–2086. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burkitt W, O’Connor G. Assessment of the repeatability and reproducibility of hydrogen/deuterium exchange mass spectrometry measurements. Rapid Commun Mass Spectrom. 2008;22:3893–3901. doi: 10.1002/rcm.3794. [DOI] [PubMed] [Google Scholar]
- 29.Tamada T, Honjo E, Maeda Y, Okamoto T, Ishibashi M, Tokunaga M, Kuroki R. Homodimeric cross-over structure of the human granulocyte colony-stimulating factor (GCSF) receptor signaling complex. Proc Natl Acad Sci U S A. 2006;103:3135–3140. doi: 10.1073/pnas.0511264103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piedmonte DM, Treuheit MJ. Formulation of Neulasta (pegfilgrastim) Adv Drug Deliv Rev. 2008;60:50–58. doi: 10.1016/j.addr.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 31.Morgan CR, Miglionico BV, Engen JR. Effects of HIV-1 Nef on human N- myristoyltransferase 1. Biochem. 2011;50:3394–3403. doi: 10.1021/bi200197e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhadra D, Bhadra S, Jain P, Jain NK. Pegnology: a review of PEG-ylated systems. Pharmazie. 2002;57:5–29. [PubMed] [Google Scholar]
- 33.Rajan RS, Li T, Aras M, Sloey C, Sutherland W, Arai H, Briddell R, Kinstler O, Lueras AM, Zhang Y, Yeghnazar H, Treuheit M, Brems DN. Modulation of protein aggregation by polyethylene glycol conjugation: GCSF as a case study. Protein Sci. 2006;15:1063–1075. doi: 10.1110/ps.052004006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu T, Li D, Manjula BN, Brenowitz M, Prabhakaran M, Acharya SA. PEGylation of Val-1(alpha) destabilizes the tetrameric structure of hemoglobin. Biochem. 2009;48:608–616. doi: 10.1021/bi801880y. [DOI] [PubMed] [Google Scholar]
- 35.Pradhananga S, Wilkinson I, Ross RJ. Pegvisomant: structure and function. J Mol Endocrinol. 2002;29:11–14. doi: 10.1677/jme.0.0290011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.