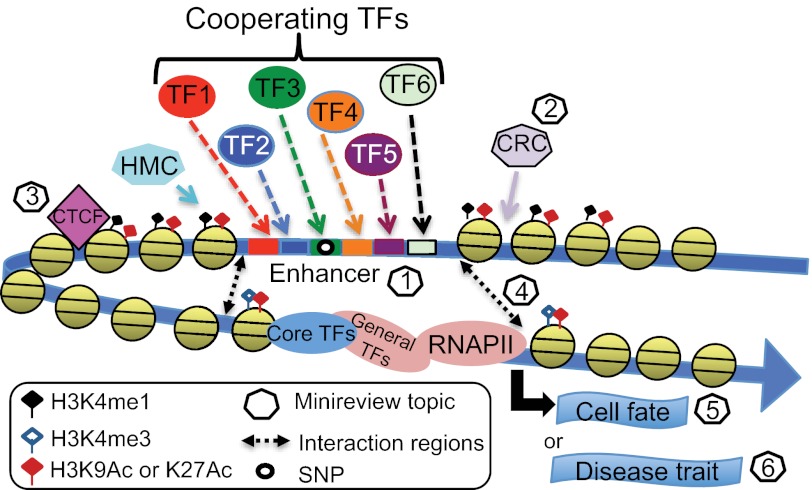
FIGURE 1.
Genome-wide characterizations of regulatory regions. Recent genome-wide ChIP-seq studies have revealed 100,00–200,000 regions of open chromatin per cell type, tens of thousands of which are marked by specifically modified histones (e.g. H3K4me1 and H3K27ac) and bound by many different site-specific transcription factors (TFs; see “Chromatin Fingerprint of Gene Enhancer Elements” by Gabriel E. Zentner and Peter C. Scacheri). Creation of these open chromatin regions requires the actions of histone-modifying complexes (HMC), chromatin-remodeling complexes (CRC), and boundary proteins such as CTCF (see “SWI/SNF Chromatin-remodeling Factors: Multiscale Analyses and Diverse Functions” by Ghia Euskirchen, Raymond K. Auerbach, and Michael Snyder and “Genome-wide Studies of CCCTC-binding Factor (CTCF) and Cohesin Provide Insight into Chromatin Structure and Regulation” by Bum-Kyu Lee and Vishwanath R. Iyer). Distal regulatory regions are thought to function by interaction of bound site-specific factors with other transcription factors bound to core promoters via looping of the intervening DNA. For a review of recent experimental and computational methods used to identify intra- and interchromosomal interactions, see “Uncovering Transcription Factor Modules Using One-dimensional and Three-dimensional Analyses” by Xun Lan, Peggy J. Farnham, and Victor X. Jin. Finally, it is becoming increasingly clear that distal regions are critically important in specifying cell fate via regulation of specific cohorts of genes (see “Transcription Factor-mediated Epigenetic Reprogramming” by Camille Sindhu, Payman Samavarchi-Tehrani, and Alexander Meissner) and that SNPs located within distal regulatory regions contribute to the development of many human diseases (see “Genome-wide Epigenetic Data Facilitate Understanding of Disease Susceptibility Association Studies” by Ross C. Hardison). RNAPII, RNA polymerase II.