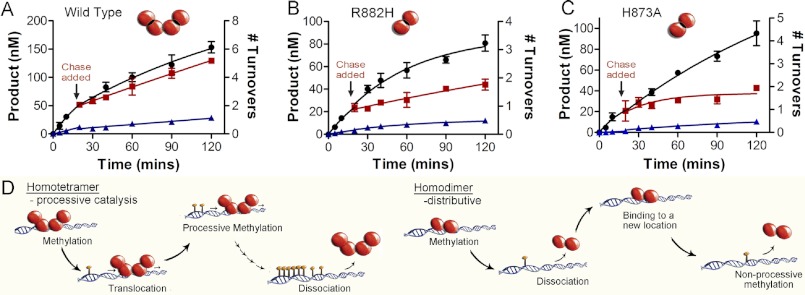
FIGURE 5.
Dimer interface disrupting mutants eliminate processive catalysis. Chase assays show DNMT3A homotetramers (WT) are processive and dimers (H873A and R882H) are non-processive, A, WT; B, H873A; and C, R882H. ●, only substrate (20 μm bp RASSF1A); ■, substrate and then 400 μm bp chase (pCpGL) at 20 min; ▴, substrate and pCpGL at the start of the reaction. Minimal methylation is detected after addition of chase DNA with the dimer mutant (H873A and R882H), unlike tetramer (WT), which shows less than 10% change in activity. D, homodimers that were formed by disrupting the dimer interface resulted in enzymes that bind DNA followed by methylation and then fast dissociation.