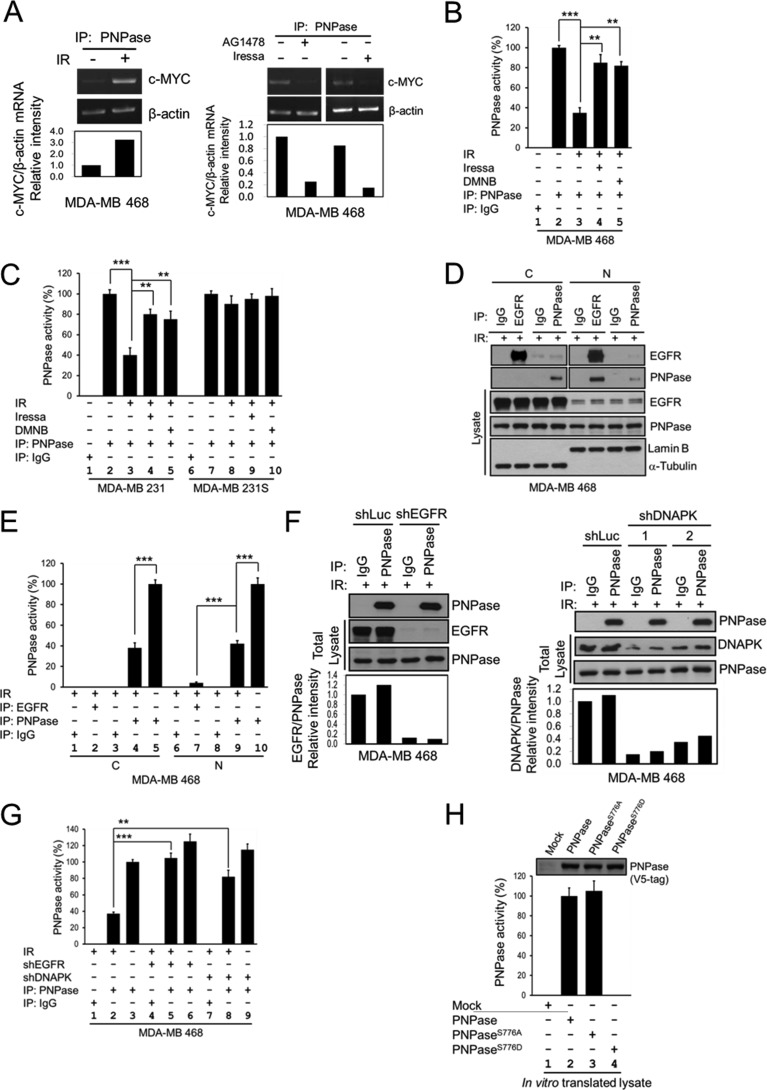
FIGURE 4.
Phosphorylation of PNPase at Ser-776 abolishes its ribonuclease activity. MDA-MB 468 cells were exposed with or without 4 Gy IR (A, left panel), or treated with or without 10 μm AG1478 or 10 μm Iressa for 12 h (A, right panel). PNPase was isolated by IP with anti-PNPase antibody from total lysates. The degradation of c-MYC mRNA after each treatment was determined by the in vitro c-MYC mRNA degradation assay with each immunoprecipitated PNPase. B, MDA-MB 468 cells were exposed with or without 4 Gy IR, or treated with or without 10 μm DMNB or 10 μm Iressa for 12 h. PNPase was isolated by individual IP with antibody against IgG or PNPase from total lysates. The ribonuclease activity of PNPase toward c-MYC mRNA after each treatment was determined by the in vitro mRNA degradation assay with each immunoprecipitated IgG or PNPase. C, MDA-MB 231 radio-resistant (231) and radio-sensitive (231S) cells were exposed with or without 4 Gy IR, or treated with or without 10 μm DMNB or 10 μm Iressa for 12 h. PNPase was isolated by individually IP with antibody against IgG or PNPase from total lysates. The ribonuclease activity of PNPase toward c-MYC mRNA after each treatment was determined by the in vitro mRNA degradation assay. D, MDA-MB 468 cells were exposed with 4 Gy IR. PNPase was isolated by individual IP with antibody against IgG or PNPase or EGFR from cytosolic (C) and nuclear (N) lysates. E, ribonuclease activity of PNPase or bound to EGFR toward c-MYC mRNA after each treatment was determined by the in vitro mRNA degradation assay by using same amount of immunoprecipitated PNPase. F, MDA-MB 468 cells were infected with lentivirus containing shRNAs against luciferase (Luc, as negative control), EGFR (shEGFR-1), or DNAPK (shDNAPK-1 and shDNAPK-2) to knock down specific gene expression. PNPase was isolated by individually IP with antibody against IgG or PNPase from total lysates. G, ribonuclease activity of PNPase toward c-MYC mRNA after each treatment was determined by the in vitro mRNA degradation assay by using same amount of immunoprecipitated PNPase. H, each PNPase variant was in vitro translated by TNT-coupled reticulocyte lysate system, and the expression of each recombinant PNPase protein was determined by IB (top). The ribonuclease activity of each in vitro translated PNPase was examined as aforementioned (lower panel). The PNPase activity was measured by the relative mRNA intensity of c-MYC/β-actin through qPCR. The activity of PNPase in normal cell culture condition or wild type from in vitro translated will be defined as 100%. ***, p < 0.005; **, p < 0.01 by t test.