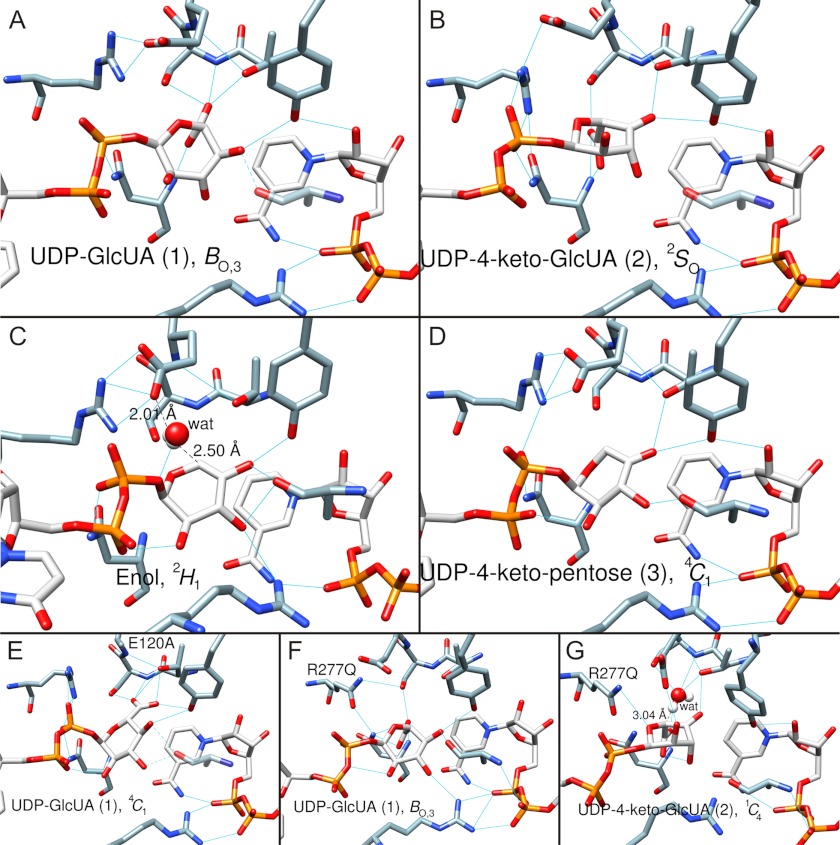
FIGURE 5.
Trajectory of sugar ring conformation along the hUXS1 reaction coordinate determined by MD simulation. Numbering of substrate/intermediate corresponds to the designation shown in Schemes 1 and 2. Blue lines indicate hydrogen bonds. Amino acid residues (except mutations) are not labeled here for better clarity (refer to Fig. 2). A, substrate UDP-GlcUA, 1, is in BO,3 boat conformation. B, UDP-4-keto-GlcUA intermediate, 2, in 2SO skew-boat conformation. C, UDP-4-keto-pentose, enol form, in 2H1 half-chair conformation. The water that protonates C5 is shown in white/red. D, UDP-4-keto-pentose intermediate, 3, in 4C1 chair conformation. E, mutant complex E120A·NAD+·UDP-GlcUA. The substrate, 1, is in the 4C1 chair conformation. F, mutant complex R277Q·NAD+·UDP-GlcUA. The substrate, 1, is in the BO,3 boat conformation. G, mutant complex R277Q·NADH·UDP-4-keto-GlcUA. The 4-keto-d-glucuronyl ring of UDP-4-keto-GlcUA, 2, is in a 1C4 chair conformation. MD simulation revealed a water molecule possibly involved in hydrogen/deuterium exchange at C5 of UDP-4-keto-GlcUA in the R277Q mutant. The water is shown in white/red. Note that the modeled structure of the corresponding NADH·UDP-4-keto-GlcUA complex of the wild-type enzyme (panel B) lacked water in a position suitable for hydrogen/deuterium exchange, consistent with only a single deuterium being incorporated during reaction in D2O.