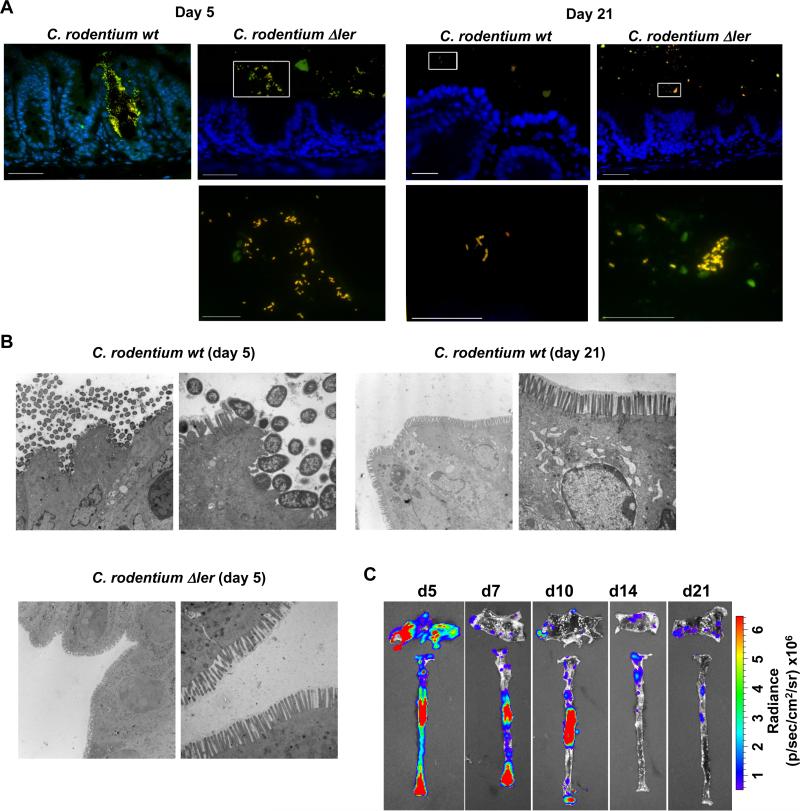
Fig. 3. Localization of C. rodentium to intestinal niches is mediated by LEE-encoded virulence factors.
A, Dual FISH staining using DNA probes that label virtually all true bacteria (EUB338, red) and the γ-Proteobacteria class to which C. rodentium belongs (GAM42a, green). Pathogenic bacteria (i.e. EUB338+/GAM42a+ cells) are yellow. Lower panels show high magnification images corresponding to boxed areas (upper panels). Note the yellow color on all the bacteria stained in the cecum infected with the Δler mutant; indicating all bacteria in the lumen are C. rodentium. Scale bar: 50μm (upper panels), 20μm (lower panels), Results are representative of 2 experiments. B, Transmission Electron Micrographs of cecum from infected GF mice at day 5 and day 21 post infection with WT C. rodentium and at day 5 with the Δler mutant. Original magnification: 3,400× (left panel) and 13,500× (right panel). Scale bar: 2 μm (left panel) and 500 nm (right panel). Results are representative of 2 experiments. C, GF mice were infected orally with WT C. rodentium carrying the ler-lux fusion. Cecum and colonic tissues were collected at the indicated day and then washed with PBS to remove non-adherent bacteria. Bioluminescent imaging of ler expression of C. rodentium attached to the cecum (top) and colon (bottom). Results are representative of 2 experiments using 4 different mice.