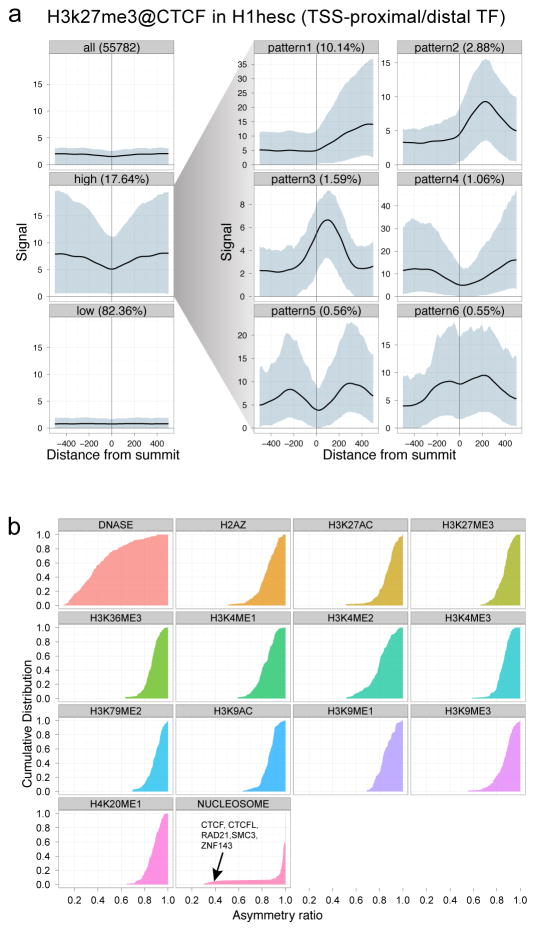
Figure 3. Patterns and Asymmetry of Chromatin Modification at Transcription Factor-binding Sites.
Panel A shows the results of clustered aggregation of H3K27me3 modification signal around CTCF binding sites (a multi-functional protein involved with chromatin structure). The first three left-most plots show the signal behaviour of the histone modification over all sites (top) and then split into the high and low signal components. The high signal component is then decomposed further into six different shape classes on the right (see ref 30 for details). The shape decomposition process is strand aware. Panel B summarises shape asymmetry for DNase1, nucleosome and histone modification signals by plotting an asymmetry ratio for each signal over all TF binding sites. All histone modifications measured in this study show predominantly asymmetric patterns at TF binding sites.