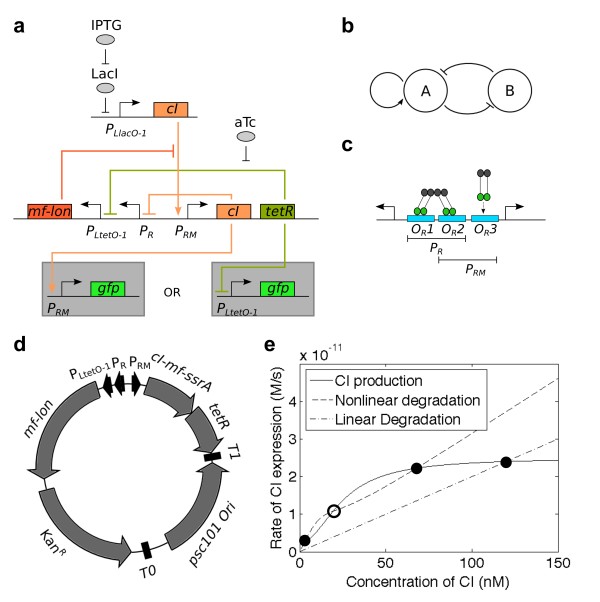
Figure 1.
Bistable switch utilizing dynamic protein degradation. (a) The hybrid transcription/enzyme bistable switch consists of two promoters and three genes. (b) The basic topology can be shown as two linked positive feedback loops. The first positive feedback look consists of the autoactivation of PRM by CI. The second positive feedback loop consists of TetR repression of PLtetO-1, and CI degradation by mf-Lon. The PR and PLtetO-1 promoters are serially placed together, but effectively act as one promoter. (c) The PRM/PR bidirectional double promoter consists of three CI dimer binding sites. CI dimers readily bind OR1 and OR2. OR3 is only bound when there are high concentrations of CI dimers. (d) The plasmid map shows that the bistable switch is built on a low copy number plasmid with pSC101 origin of replication and kanamycin marker. (e) The rate balance plot shows the difference between nonlinear degradation vs. standard linear degradation. The steady-state solutions to the concentrations of CI (and thus, system state) arise at the intersection of the CI production curve and either of the degradation curves. A linear degradation curve produces one steady-state solution. Nonlinear degradation creates three intersects with the CI production line to produce a bi-stable system. Solid circles indicate stable steady-states. Open circles represent unstable steady-states.