Figure 1. 3D-SIM images of FtsZ-GFP localization in live cells of B. subtilis.
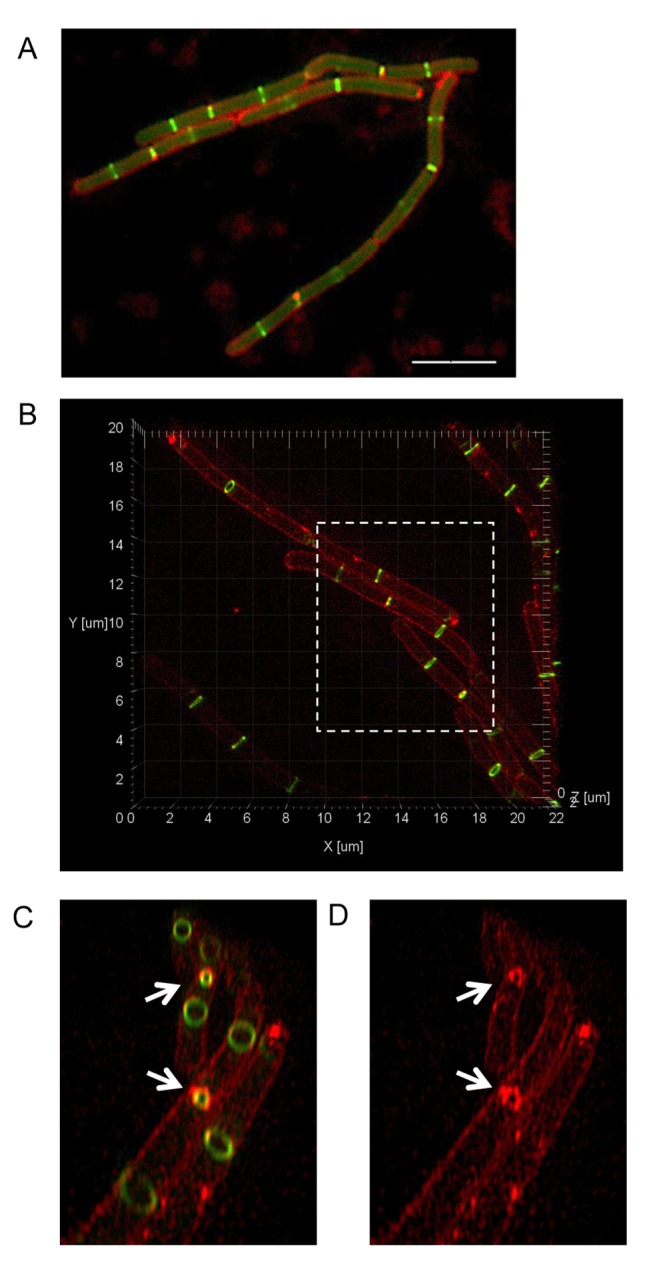
(A) Conventional wide-field fluorescence microscopy (Zeiss) image of B. subtilis strain SU570 (ftsZ-gfp) stained with the membrane dye FM4-64 shows how FtsZ-GFP assembles into Z rings. Scale bar, 5 µm. (B) When the same strain is imaged using 3D-SIM (OMX V3), regions of interest from the image can be selected (dashed box) to zoom in and rotate the image around the z-axis to view 3D FtsZ structures in the axial plane. (C–D) The improved image resolution provided by 3D-SIM allows visualization of constricting Z rings and the inner the cell membrane during division (indicated by white arrows).
