Figure 7. EzrA rings in S. aureus are also heterogeneous and show dynamic movement.
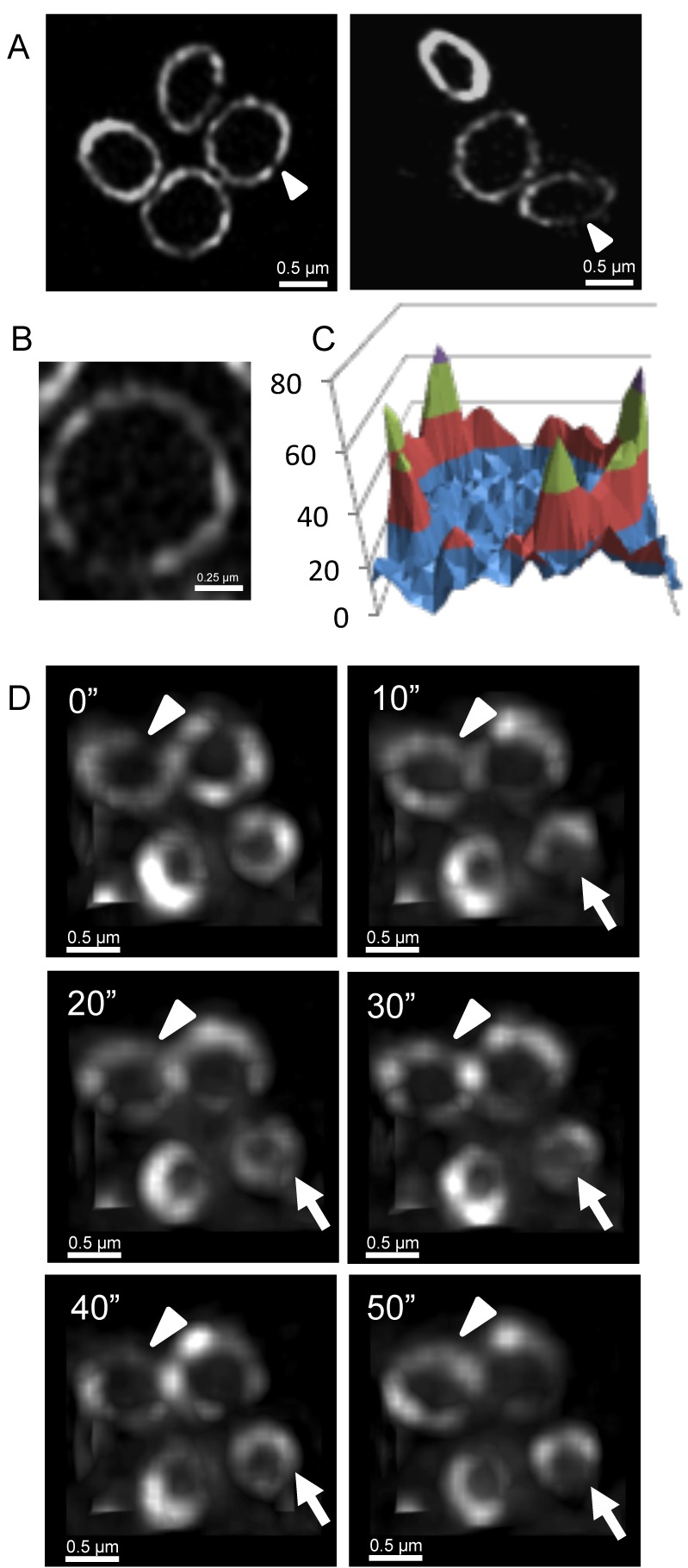
(A) To examine EzrA-GFP localization using 3D-SIM (OMX V3), S. aureus strain SA126 was grown in L-broth at 37°C. The EzrA-GFP fusion protein displays a similar type of localization at the division site as FtsZ. (B) A close-up image of an EzrA ring in SA126 cells imaged in the lateral orientation with 3D-SIM (OMX V3). (C) A 3D intensity plot of EzrA rings orientated in the lateral plane shows a similar profile to FtsZ-GFP. (D) Deconvolved images were obtained through conventional wide-field fluorescence to monitor the localization of EzrA-GFP over time (acquired using OMX Blaze system). EzrA-GFP localization is dynamic and similar to that observed using FtsZ-GFP in S. aureus. White arrowheads show areas where the EzrA concentration is reduced. Arrowheads indicate the formation of additional gaps in the EzrA-GFP rings. Time (s) is indicated on the upper left-hand side for each image.
