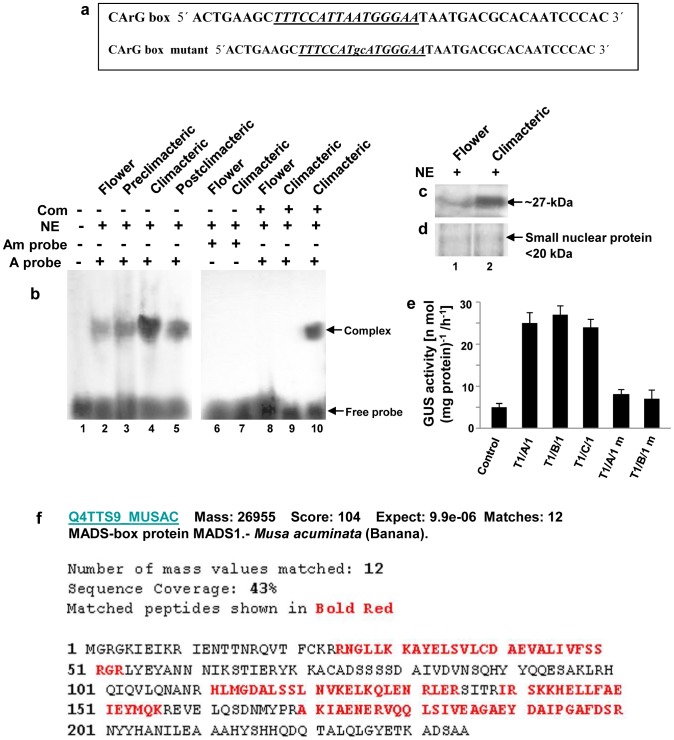
Figure 1. Detection of CArG-box DNA binding protein in banana fruit and flower nuclear extracts.
a CArG box motif, derived from Arabidopsis Agamous MADS box binding site, was used as probe for gel mobility shift assays. The MADS domain consensus binding site (CArG box motif) has been indicated (italics and underlined). DNA fragment containing the modified CArG box motif (highly conserved T and A residues were changed to G and C residues) has been indicated (small letters underlined). b Gel mobility shift assay using labeled CArG-box DNA as probe (A probe). Lane 1 contained only radiolabeled CArG-box DNA probe, while 15 µg nuclear protein extract was added in lanes 2–10. Lane 2 to 5 contained nuclear protein extract from banana flower and fruit pulp tissues at the preclimacteric, climacteric and postclimacteric stages of ripening with labeled CArG box motif as probe. Lane 6 and 7 contained nuclear protein extract from banana flower and climacteric fruit pulp with labeled modified CArG box motif as probe (Am probe). In lane 8 and 9, nuclear protein extract from banana flower and climacteric pulp was incubated with labeled CArG box motif in presence of 100-fold excess of unlabeled (non-radioactive) CArG box motif. Lane 10 contained nuclear protein extract from climacteric banana pulp with labeled CArG box motif in presence of 100-fold excess of unlabeled modified CArG box motif. Com-competitor, NE-nuclear extract. c South-Western blot analysis with nuclear protein extracts isolated from flower and climacteric banana fruit pulp (lanes 1–2). ∼25 µg of nuclear extract was loaded in each lane. The radiolabeled synthetic oligo containing CArG box motif was used as probe. d Equal amounts of small nuclear protein (SNP) from banana flower and climacteric pulp tissues were resolved in 12% SDS-PAGE and has been shown as loading control. e Measurements of GUS activity in transgenic tobacco lines carrying trimeric CArG-box motif (3X CArG) or the modified trimeric CArG-box sequence (m) in fusion with GUS. GUS activity was detected in the leaves of control and transgenic tobacco lines. The error bars indicate mean values from three independent observations. f Identification of 27-kDa CArG-box binding MADS-domain protein by mass spectrometry. Overall sequence coverage of the peptides with the matched protein (Q4TTS9_MUSAC of Musa acuminata). Matched peptides shown in red letters. Experiments were repeated three times. Representative images from at least three independent experiments are shown for Figure b–d.