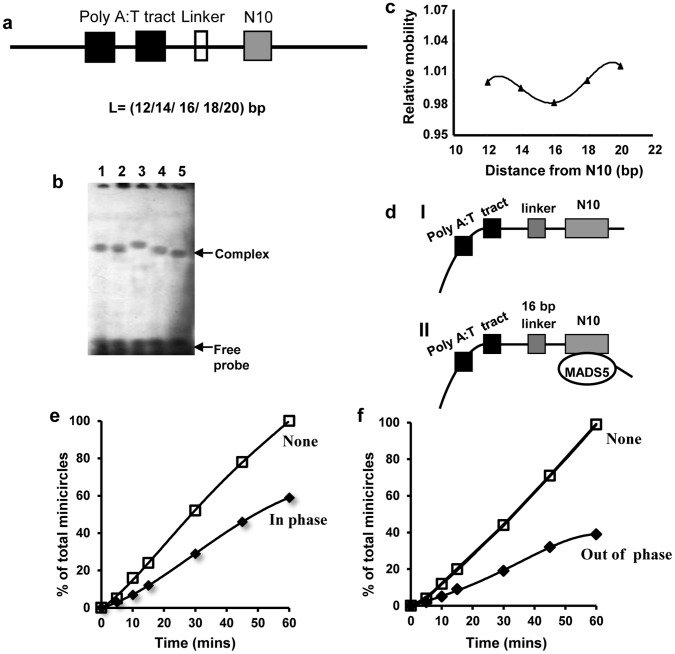
Figure 4. Phasing analysis of MA-MADS5 induced DNA bending in complexes with N10-site. a.
Diagrammatic representation of phasing probes. The sequence giving rise to the intrinsic bend (filled box) is composed of two helically phased hexameric [A:T] tracts. The linker lengths (open box) varies between 12 to 20 bp, giving rise to spacing of 51 and 59 bp respectively between the centre of the first hexameric [A:T] tract and the centre of the CArG box (N10-site) (grey box). b Bacterially expressed recombinant MA-MADS5 protein was incubated with each of the phasing probe and analyzed by electrophoresis through a 6% non-denaturing polyacrylamide gel. The mobilities of free probes have been indicated. The different linkers in each probe resulted in spacer lengths of 51 bp (lane 1), 53 bp (lane 2), 55 bp (lane 3), 57 bp (lane 4) and 59 bp (lane 5) respectively between the centre of the first hexameric poly [A:T] tract and the centre of the CArG box (N10-site). c The relative motilities of DNA-protein complexes were normalized for differences in probe mobilities and plotted as a function of the distance between the centre of the N10 from the intrinsic DNA bend. d Diagrammatic representation of the structure of the complexes containing an intrinsic DNA bend (I), MA-MADS5 induced bend on N10-site and ‘in-phase’ (II) with the intrinsic DNA bend. e–f Ligase mediated circularization analysis of MA-MADS5 protein on the N10 site. Assays were carried out on sites containing the linker of 16 and 20 bp in which the centre of the N10 site was ‘in-phase’ (e) or ‘out-of-phase’ (f) with respect to the centre of a first hexameric poly [A:T] tract. Minicircle formation after incubation with DNA ligase for the indicated time points (0 to 60 min) has been graphically represented. The circularization reactions were carried out in the absence or presence of MA-MADS5 protein. Experiments were repeated three times.