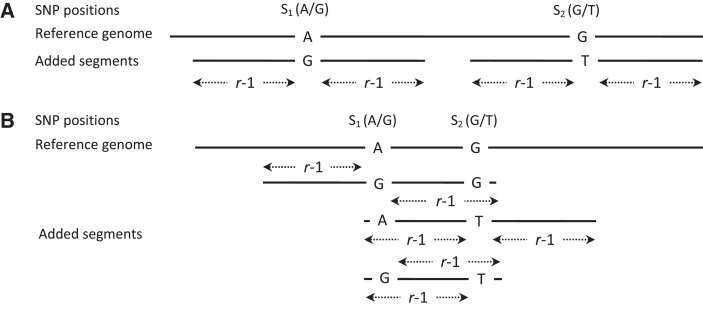
Figure 1.
Schematic representation of the enhanced segments added for two SNPs, S1 and S2. The read length is indicated by r. (A) Enhanced segments added when the distance between two adjacent SNPs S1 and S2 is ≥r. No read can overlap both SNPs in this scenario. A single enhanced segment with the non-reference allele is added for each SNP. The enhanced segment extends r − 1 bases on either side of the SNP to ensure an exact match with any read carrying the non-reference allele. (B) Scenario when the distance between S1 and S2 is <r − 1. Because there can be reads that overlap both SNPs, we need to add three segments to cover all possible haplotypes formed by S1 and S2. It is also necessary that none of the added segments is identical to another enhanced segment (or the reference) in any window of length ≥r. This ensures that the read uniquely maps to the reference or one of the enhanced segments. Multiple solutions satisfying these conditions are possible. The figure shows one such possible solution.