Abstract
Folding mechanisms in which secondary structures are stabilized through the formation of tertiary interactions are well documented in protein folding but challenge the folding hierarchy normally assumed for RNA. However, it is increasingly clear that RNA could fold by a similar mechanism. P5abc, a small independently folding tertiary domain of the Tetrahymena thermophila group I ribozyme, is known to fold by a secondary structure rearrangement involving helix P5c. However, the extent of this rearrangement and the precise stage of folding that triggers it are unknown. We use experiments and simulations to show that the P5c helix switches to the native secondary structure late in the folding pathway and is directly coupled to the formation of tertiary interactions in the A-rich bulge. P5c mutations show that the switch in P5c is not rate-determining and suggest that non-native interactions in P5c aid folding rather than impede it. Our study illustrates that despite significant differences in the building blocks of proteins and RNA, there may be common ways in which they self-assemble.
INTRODUCTION
In RNA folding, the stability gap separating the free energy of the native state from other non-native structures is often small (1–3). Thus, misfolded conformations in which even the secondary structure contains errors can form with substantial probability (4). One way to correct errors in the secondary structure is by coupling base pair reorganization with RNA tertiary folding, either by realigning entire helices (5–7) or by shuffling non-Watson–Crick base pairs within a helix (8,9). Although it has been known for some time that certain base pairs are not stable until the RNA tertiary structure forms (10), tertiary interactions may also be used to correct non-native secondary structures that are present in the RNA. Direct coupling between RNA secondary and tertiary structure challenges the conventional view (3,11) that native secondary structures always form before the establishment of tertiary interactions.
The ease with which RNAs form alternative stable structures raises the possibility that tertiary interaction-driven base pair switching, and not merely helix stabilization, might be more widespread than commonly assumed. To explore this possibility, we investigated the refolding kinetics of the P5abc RNA that switches its secondary structure when inter-helix tertiary interactions are formed (Figure 1) (6,12). The P5abc RNA is an independently folding tertiary domain of the Tetrahymena thermophila group I ribozyme (13,14) which is one of the fastest regions of the ribozyme tertiary structure to form (15). P5abc is composed of three helices (paired regions P5a, P5b and P5c) that interact via a rich web of tertiary contacts in the folded state (16). One set of tertiary interactions is formed between an A-rich bulge in helix P5a and the loop of helix P5c and another is formed by tandem G-A pairs at the junction between helices P5a, P5b and P5c.
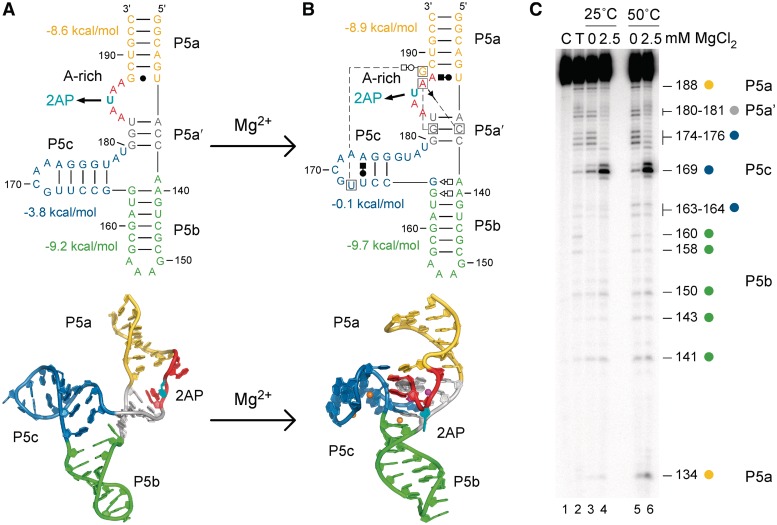
Figure 1.
Structures of the tP5abc RNA. (A) The secondary structures (top) and ribbon diagrams (bottom) of the extended tP5abc from NMR (6) are compared with (B) the folded P5abc structure from X-ray crystallography (22). Mg2+ ions visible in the X-ray structure are represented by spheres. Nucleotides are numbered discontinuously because 5 bp of the natural P5b are missing in tP5abc. Non-Watson–Crick base pairs are represented by Leontis/Westhof symbols (49). U185 was replaced with the fluorescent analog 2AP (cyan), which unstacks in the folded RNA. Base-pairing free energies are from mfold (35). (C) RNase T1 probing of tP5abc secondary structure at 25 and 50°C, with no Mg2+ (extended) or 2.5 mM Mg2+ (folded). Lane C, no digestion control; lane T, RNase T1 sequencing ladder.
A nuclear magnetic resonance (NMR) structure of a truncated form of the P5abc domain (tP5abc) showed that the P5abc secondary structure in the absence of Mg2+ (Figure 1A) differs significantly from that of the Mg2+-bound state (Figure 1B), implying that tertiary folding is coupled to a reorganization of the RNA secondary structure (6). First, the base pairs in P5c change register, expanding the stable GNRA tetraloop in the extended form of P5c to a less stable pentaloop in the native state that positions U168 to interact with G188 in P5a (6,12,17). Second, reorganization of the three-helix junction replaces base pairs A139-U179 and G164-U177 with tandem G-A pairs (A139-G164 and A140-G163) in the native from of P5b.
Although the native secondary structure of P5abc is less stable than its extended secondary structure, this is compensated by the favorable free energy of tertiary interactions in the presence of Mg2+ ions (17,18). In addition to non-specific charge stabilization (19,20), Mg2+ ions specifically coordinate a ‘corkscrew’ in the backbone of the A-rich bulge (magenta spheres; Figure 1B), while four additional ions interact with P5c and with tandem G-A pairs in P5b (orange spheres; Figure 1B) (21,22). Evidence for specific binding of Mg2+ to P5abc RNA in solution comes from NMR, phosphorothioate substitution-interference (12,21) and refolding experiments (23,24).
Force denaturation experiments confirmed the importance of tertiary interactions for global stability of the P5abc domain, as high forces were required to rupture the P5abc structure in Mg2+ (25). More importantly, when the force was relaxed, refolding occurred in two steps by a kinetic partitioning mechanism (26). Of particular relevance is that the single-molecule experiments showed that folding of P5abc, upon release of force, involves intermediates regardless of the Mg2+ concentration. However, such experiments provide no information about the structure of the intermediates nor their location along the refolding pathways.
Herein, we provide evidence that helix switching occurs in the same step of folding as the formation of tertiary interactions in P5abc. Using a fluorescence assay, we show that the solution refolding pathway contains at least one intermediate. However, experiments and simulations show that the base pair shift in P5c does not limit the rate of folding. Instead, our results show that switching of the P5c helix occurs in the same folding step as the formation of tertiary interactions in the A-rich bulge and that both of these events follow the slow step of folding. Our results show that the energy landscape of even the rapidly folding P5abc subdomain is rugged, exhibiting many of the complexities found in the folding kinetics of larger ribozymes.
MATERIALS AND METHODS
Coarse-grained dynamics simulations
Coarse-grained simulations used the three interaction site (TIS) model (Supplementary Figure S1) in which each nucleotide is represented by three interactions sites corresponding to the centers of masses of the base, sugar and phosphate (27). The force field for the TIS model includes base-stacking interactions which were taken from empirical energy parameters (28). G•A pairs were given the same weight as A-U pairs. Replacing G•A parameters with those for G-C pairs resulted in negligible differences to our simulation results (compare Figure 3 and Supplementary Figure S2).
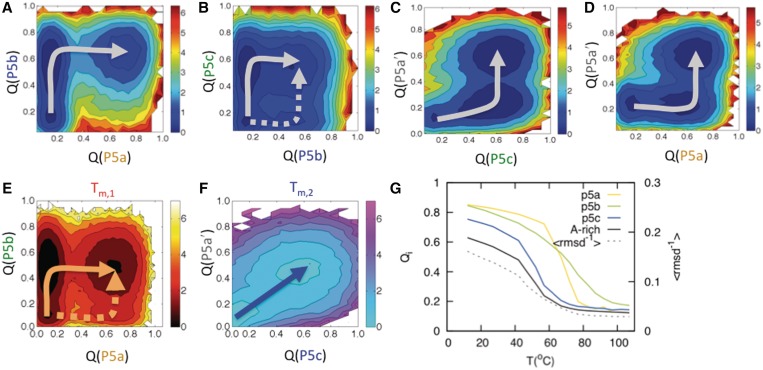
Figure 3.
Free energy landscapes of tP5abc from simulation. Free energy profiles were projected to the fraction of native contacts (Q) for each region of tP5abc as defined in Figure 1. The free energy profile is calculated at the melting temperature in units of kBT. (A–D) Energy profiles for the extended RNA. (E and F) Energy profiles for the folded state, at the high temperature transition (E) or low temperature transition (F). (G) Comparison of secondary structures (Qi) with native tertiary contacts, as reported by the overall root mean square deviation (dashed line).
For thermodynamic analyses, multiple trajectories containing numerous folding and unfolding transitions were collected. All the simulations were performed using Langevin dynamics in the low friction limit (ζ = 0.05 τL−1), which enhances the rate of conformational sampling (29). The unit of time is 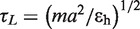 , in which the mass of the RNA beads is m = 0.1–0.16 kg/mol, the average distance between beads is a = 4.6 Å and the energy scale is εh ∼1 kcal/mol. Profiles of the specific heat with respect to temperature, Cv(T), were obtained using Weighted Histogram Analysis Method (WHAM), and a peak in Cv(T) designates a melting transition temperature. The trajectories at the melting temperature were collected to generate the free energy profile with respect to the reaction coordinate, Q, the fraction of native contacts.
, in which the mass of the RNA beads is m = 0.1–0.16 kg/mol, the average distance between beads is a = 4.6 Å and the energy scale is εh ∼1 kcal/mol. Profiles of the specific heat with respect to temperature, Cv(T), were obtained using Weighted Histogram Analysis Method (WHAM), and a peak in Cv(T) designates a melting transition temperature. The trajectories at the melting temperature were collected to generate the free energy profile with respect to the reaction coordinate, Q, the fraction of native contacts.
RNA preparation
Truncated P5abc RNA (6) containing 2-aminopurine (2AP) was chemically synthesized (Dharmacon) and purified by denaturing 10% polyacrylamide gel electrophoresis. The RNA solution was desalted and concentrated by ultracentrifugation (Centricon Plus-20; Millipore) and stored at −20°C in water. The typical yield after purification was 10–20%.
UV thermal denaturation
Thermal denaturation of P5abc was monitored at 260 nm at a heating rate of 0.1°C/min on a Cary 400 spectrophotometer. tP5abc RNA (2 µM) was dissolved in 10 mM Na–cacodylate (pH 7.5), 40 mM NaCl plus the desired concentration of MgCl2. Global Melt Fit provided by D. Draper (http://ded.chm.jhu.edu:16080/~draper/) was used to obtain enthalpies and melting temperatures, assuming two transitions and temperature-dependent baselines.
RNase T1 partial digestion
5′-[32P]-labeled tP5abc was prepared with T4 polynucleotide kinase and gel-purified as described previously (30). The RNA was equilibrated 10 min in CE buffer (10 mM cacodylate, pH 7.5; 0.5 mM EDTA, pH 8.5) with or without 2.5 mM MgCl2 before treatment with 0.1 U RNase T1. Products were resolved on a 12% sequencing gel.
Equilibrium Mg2+ titrations
For equilibrium folding, 2 ml 0.5 µM RNA in CE and 2 ml 0.5 µM RNA in CE plus MgCl2 were denatured 5 min at 50°C and degassed. These solutions were mixed to produce Mg2+ titrations and incubated 5 min at 30°C after each addition before the emission spectra (3–5 scans) were recorded with excitation at 310 nm (Perkin Elmer LS50B). Excitation and emission slit widths were 15 and 10 nm, respectively.
The fluorescence intensity F at 370 nm versus Mg2+ concentration, C, was fit to
| (1) |
in which Cm is the midpoint of the folding transition, n is the Hill coefficient, αU is the fluorescence intensity at zero Mg2+ and a linear baseline for the folded RNA is given by αN and βN. The ΔG of folding was calculated from 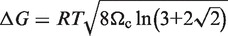 , in which
, in which 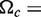
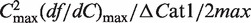 is a measure of the cooperativity (31). Unfolding by dilution of Mg2+ produced similar results (data not shown).
is a measure of the cooperativity (31). Unfolding by dilution of Mg2+ produced similar results (data not shown).
Urea denaturation
At equilibrium, 0.5 µM RNA in CE plus 1 mM MgCl2 was mixed with the same solution also containing 9 M urea to produce the desired final urea concentration. The fluorescence data were fit to the linear binding model (32,33):
| (2) |
in which D is the concentration of urea, Dm is the concentration of denaturant at the midpoint of the unfolding transition and m is a constant that reflects the change in folding free energy with denaturant concentration. Linear baselines (α + βD) corrected for a slight change in the fluorescence of the folded state with urea, and a larger increase in 2AP fluorescence of the extended state with urea as stacking between 2AP and the adjacent bases was disrupted. The data were normalized as in Equation (1).
Folding kinetics
Folding kinetics was measured on an Applied Photophysics SX.18MV stopped-flow spectrometer with a dead time of 1.3 ms. The excitation wavelength was 310 nm, and the emission signal was measured using a 360 nm wide band cut-off filter. Folding reactions were performed by mixing tP5abc RNA prepared as mentioned earlier (1 µM in CE) with an equal volume of CE plus 2 × MgCl2. Unfolding reactions were performed by pre-folding the RNA in CE plus 1 mM MgCl2 and diluting it 26-fold with buffer to reach the desired final MgCl2 concentration. Unfolding rates in urea were obtained by mixing 1 volume folded RNA (CE, 1 mM MgCl2) with 10 volumes urea in CE, 1 mM MgCl2. Folding rates were obtained by diluting RNA in 5.5 M urea 10:1 with buffer. Individual kinetic traces were fit to a single exponential rate equation and the resulting rate constants averaged, except for folding reactions of G174A, C166U RNA which were fit to a double exponential equation.
The observed rate kobs as a function of Mg2+ concentration C was fit by a three-state model 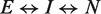 , in which the forward rate was given by kobs,f = kfCr, where r is the apparent number of ions linked to the slow step of the forward folding reaction (7). The unfolding pathway (N to I) was assumed to depend on the stability of N. Expressions for the forward and reverse paths were combined yielding the following equation:
, in which the forward rate was given by kobs,f = kfCr, where r is the apparent number of ions linked to the slow step of the forward folding reaction (7). The unfolding pathway (N to I) was assumed to depend on the stability of N. Expressions for the forward and reverse paths were combined yielding the following equation:
| (3) |
in which kf and ku are the folding and unfolding rate constants, K2 is the midpoint and m is the slope of the equilibrium between I and N.
β values
The values of β for the Mg2+ titrations were calculated from β = n‡/nH, in which nH = dlnK/dlnC was taken from the equilibrium folding curves and n‡ = dlnkobs/dlnC was taken from the folding kinetics over the Mg2+ concentration ranges in which the folding and unfolding arms were linear.
RESULTS
Thermal denaturation
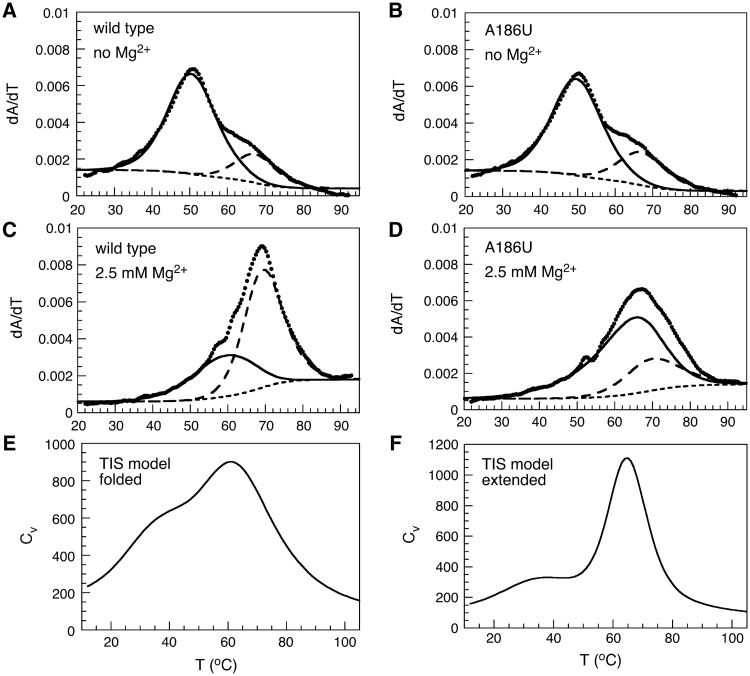
For these studies, we used the 56 nt tP5abc, which lacks five base pairs in the center of P5b and three nucleotides at the top of P5a (6) (Figure 1) yet forms the correct tertiary structure around the three-helix junction (12,17). UV thermal denaturation experiments in the absence of Mg2+ showed that the extended secondary structure unfolds in two transitions (Figure 2A) with midpoints at 50 and 68°C (Supplementary Table S1). The melting profile in the absence of Mg2+ was altered very little by the mutation A186U, which disrupts tertiary interactions between the A-rich bulge and P5c but does not change the predicted secondary structure (Figure 2B). In 2.5 mM Mg2+, the secondary structure opened at a higher Tm, as expected (Figure 2C and D). Moreover, the ‘wild type’ tP5abc exhibited an additional low temperature unfolding transition not visible for the A186U mutant, which we assigned to denaturation of the tertiary structure.
Figure 2.
Thermal stability of tP5abc RNA. (A–D) The first derivative of absorbance at 260 nm with temperature was fit to two transitions. All experiments were in 40 mM NaCl, 10 mM Na-cacodylate (pH 7.5). Symbols: dotted lines, data; solid lines, first transition; long dashed lines, second transition; short dashed lines, baseline. (E and F) Simulated thermal denaturation profiles showing the specific heat Cv versus temperature for the folded (E) and extended (F) structures of tP5abc.
RNase T1 probing
We next probed the tP5abc secondary structure by RNase T1 partial digestion at 25 and 50°C to verify that Mg2+ induces the expected shift in P5c base pairing (Figure 1C). At 25°C, which is below the first thermal transition, Gs in P5a, P5b and at the junction between P5b and P5c (G163 and G164) were protected from cleavage, indicating that these helices are stable at 25°C in the extended structure (lane 3, Figure 1C). As expected, these residues were cleaved more strongly at 50°C, the Tm of the first thermal unfolding transition (lane 5, Figure 1C).
When the tP5abc was folded in 2.5 mM MgCl2, G174-G176 in P5c and G180-G181 in P5a′ also became protected at 25°C, showing that these base pairs near the three-helix junction become more stable in the tertiary structure (lane 4, Figure 1C). Although bases in P5c are predicted to be paired in the extended structure, this helix may breathe or rearrange more than expected in low salt conditions. However, Mg2+ strongly enhanced cleavage of G169 in the loop of P5c, confirming the expected shift from a sheared G-A pair in the tetraloop of the extended form to an unpaired and exposed G in the pentaloop of folded tP5abc (lanes 4 and 6, Figure 1C).
Simulations of structural transitions during thermal unfolding
To interpret the unfolding pathway implied by the thermal denaturation curves, we performed coarse-grained molecular dynamics simulations of tP5abc using the TIS model in which each nucleotide is represented by three beads (27). We performed two sets of simulations, in which the TIS Hamiltonian was derived from either the extended tP5abc structure or the folded tP5abc structure (Supplementary Figure S1). In simulations of folded tP5abc, the specific heat (Cv) profile had a well-defined shoulder (Figure 2E), indicating two distinct melting transition temperatures as observed in our experiments (Figure 2C). In contrast, the Cv profile for the extended tP5abc had one peak (Figure 2F), corresponding to a single melting transition very similar to that of the A186U mutant in the presence of Mg2+ (Figure 2D). The overall specific heat profiles were in good agreement with the measured profiles, with similar transition temperatures, relative peak heights and overall shape.
To quantify the structural transitions associated with the melting of tP5abc, we calculated a series of 2D, temperature-dependent free energy profiles, F(Qi, Qj), in which Qi and Qj are the fractions of native contacts of the individual helices in the extended tP5abc or folded tP5abc (Figure 3). Although the single peak in the Cv profile indicated cooperative folding of extended tP5abc (Figure 2F), the free energy profiles revealed intermediates corresponding to the formation of individual helices. The 2D landscapes showed that the internal helices P5b and P5c form independently of each other with similar free energy barriers, in agreement with the RNase T1 data, while P5b is prerequisite for the closing helix, P5a (Figure 3A and B). P5a and P5c in turn stabilize P5a′, completing the extended secondary structure (Figure 3C and D). Because the extended form of P5c is much more stable than the native P5c helix (Figure 1A and B), our simulations predict that it forms early in the folding while native P5c does not.
To compare the folding mechanism of the extended structure to that of the folded structure, we constructed free energy landscapes for folded tP5abc at each of the two melting temperatures. These landscapes showed that the high temperature transition corresponds to sequential unfolding of P5a then P5b (Figure 3E), while the lower melting temperature corresponds to the cooperative unfolding of P5c and P5a′ (Figure 3F). Thus, under native conditions, P5c and P5a′ adjacent to the A-rich bulge fold cooperatively, rather than sequentially as in the extended state.
We next compared formation of tertiary structure, represented by the overall root mean square deviation, with secondary structure formation as measured by Qi (Figure 3G), and found that the tertiary structure forms in the lower temperature transition, concurrently with P5c and P5a′. Overall, the experimental data and the simulations show that native tP5abc unfolds through at least one equilibrium intermediate, corresponding to the loss of tertiary interactions and P5c. Our finding that the native P5c helix forms (or unfolds) in the same thermal transition as the tertiary structure shows that these steps are strongly coupled as previously suggested (6) and in agreement with the experimental RNase T1 results (Figure 1C).
Folding P5abc in Mg2+
We next asked whether similar intermediates were populated when P5abc refolds in solution. To study the Mg2+-dependent folding pathway of tP5abc experimentally, we introduced the fluorescent base analog 2AP at position 185 (cyan; Figure 1A). U185 stacks with adjacent adenosines in the extended conformer but flips out in the native structure. When U185 was replaced by 2AP, the fluorescence intensity of the labeled RNA increased upon titration with Mg2+, consistent with unstacking of this base as the RNA folds (Supplementary Figure S3A).
Several control experiments indicated that the structure of tP5abc was not significantly perturbed by the 2AP substitution. First, the 2AP substitution made almost no difference to the thermal denaturation profiles of the RNA (Supplementary Figure S3B). Second, the folding time (t1/2 = 180 ms in 0.5 mM MgCl2 at 30°C; Supplementary Figure S3C) was compatible with previous estimates of the folding kinetics from time-resolved hydroxyl radical footprinting (34) and NMR (12). Finally, a similar Mg2+-dependent folding transition was observed when pyrene was covalently attached to the 2′OH of U142 (Supplementary Figure S4).
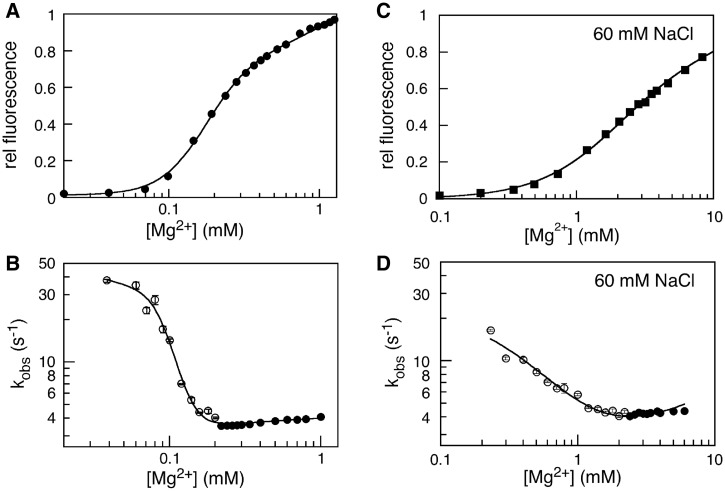
The stability of tP5abc was obtained from the increase in fluorescence intensity with Mg2+ concentration at equilibrium (Figure 4A). The midpoint of the folding transition (Cm) was 0.18 mM Mg2+ at 30°C (in 20 mM Na-cacodylate), reasonably similar to previous results (17). From the steepness of the folding transition with respect to Mg2+ concentration (31), we obtained ΔGUN = −1.3 ± 0.5 kcal/mol (Supplementary Table S2).
Figure 4.
Mg2+ dependence of folding at 30°C. (A) Cooperative change in the folding equilibrium with Mg2+. The fluorescence intensity at 370 nm versus Mg2+ concentration in 10 mM CE was fit to a cooperative model [Equation (1)]; see Supplementary Table S2 for fit parameters. (B) Observed folding kinetics versus Mg2+ concentration. Errors are the standard deviation of three experiments. The line represents the best fit to the model in Equation (3), with ku = 36 s−1, kf = 4 s−1 (Supplementary Table S3). Filled and open symbols represent kinetics from folding and unfolding reactions, respectively (see ‘Materials and Methods’ section). (C) The midpoint of the Mg2+ titration was 10-fold higher in 60 mM Na+ than in CE buffer alone because the Na+ ions compete with Mg2+ for access to the RNA (Cm = 1.8 mM; Supplementary Table S2). (D) Observed folding rate versus Mg2+ concentration in 60 mM NaCl, as in (B), with ku ≈ 40 s−1 and kf = 3.6 s−1 (Supplementary Table S3).
Mg2+ dependence of the folding kinetics
To probe the folding pathway of tP5abc in solution, the folding and unfolding kinetics was measured by stopped-flow fluorescence in Mg2+ concentrations spanning the equilibrium transition (Figure 4B). The observed unfolding rate became faster as the Mg2+ was reduced below 0.2 mM, which was expected because the equilibrium no longer favors the folded state (Figure 4B). Surprisingly, however, the observed folding rate did not increase as the Mg2+ was raised above 0.2 mM, although the native tertiary structure was becoming more stable. These results showed that the folding rate is determined by a slow step that does not depend on the stability of the native state or the recruitment of additional Mg2+ ions to the RNA. Higher salt did not eliminate the Mg2+-independent slow step in the folding pathway, as similar results were obtained when the tP5abc folding kinetics were measured in the presence of 60 mM NaCl (Figure 4C and D and Supplementary Table S3).
Structural reorganization during folding
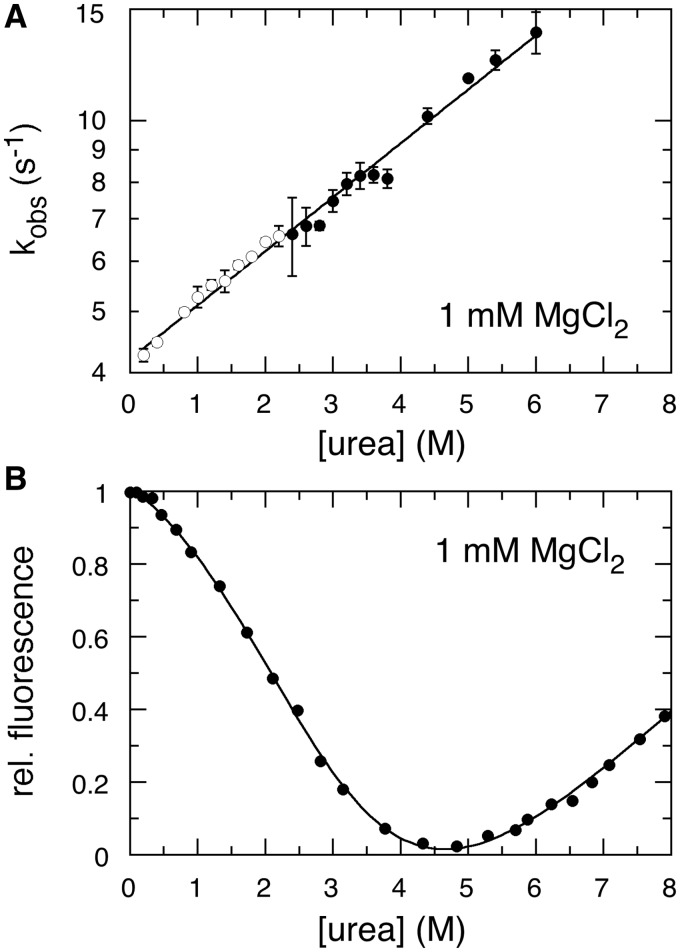
To estimate how many interactions must reorganize during folding, we used an Arrhenius plot to measure the activation energy for folding in 0.5 mM Mg2+, which was 21.3 kcal/mol (Supplementary Figure S5). This activation energy is comparable to ΔH for opening five base pairs based on empirical free energy parameters (35). Next, we measured the folding equilibrium and kinetics of tP5abc in urea. In 1 mM Mg2+, urea denaturation of native tP5abc at equilibrium had a slope of m = 0.52 kcal/mol•M (Figure 5A). Based on studies of RNA duplexes and tRNA, this sensitivity to urea is comparable to the loss of four to five base pairs or the exposure of 2900 Å2 of buried surface (33). Strikingly, urea strongly increased the folding rate under both native and denaturing conditions (Figure 5B). From these results, we concluded that tP5abc refolds through one or more intermediates and that formation of the intermediate requires the disruption of interactions in the extended state. However, we show below that this reorganization is not due to opening of P5c.
Figure 5.
Urea increases the folding rate. (A) Urea denaturation of tP5abc in 1 mM Mg2+ at 30°C, fit to Equation (2). The increase in fluorescence intensity between 5 and 8 M urea is due to denaturation of the secondary structure and the loss of stacking interactions between 2AP and adjacent bases. (B) Observed folding rate at 30°C increases with urea concentration. Errors are the standard deviation for three experiments. Unfolding rates (open circle) were obtained by mixing prefolded RNA (1 mM Mg2+) with urea in CE, 1 mM MgCl2. Folding rates (filled circle) were obtained by diluting denatured RNA in 5.5 M urea with CE plus 1 mM Mg2+.
Switching of the P5c helix
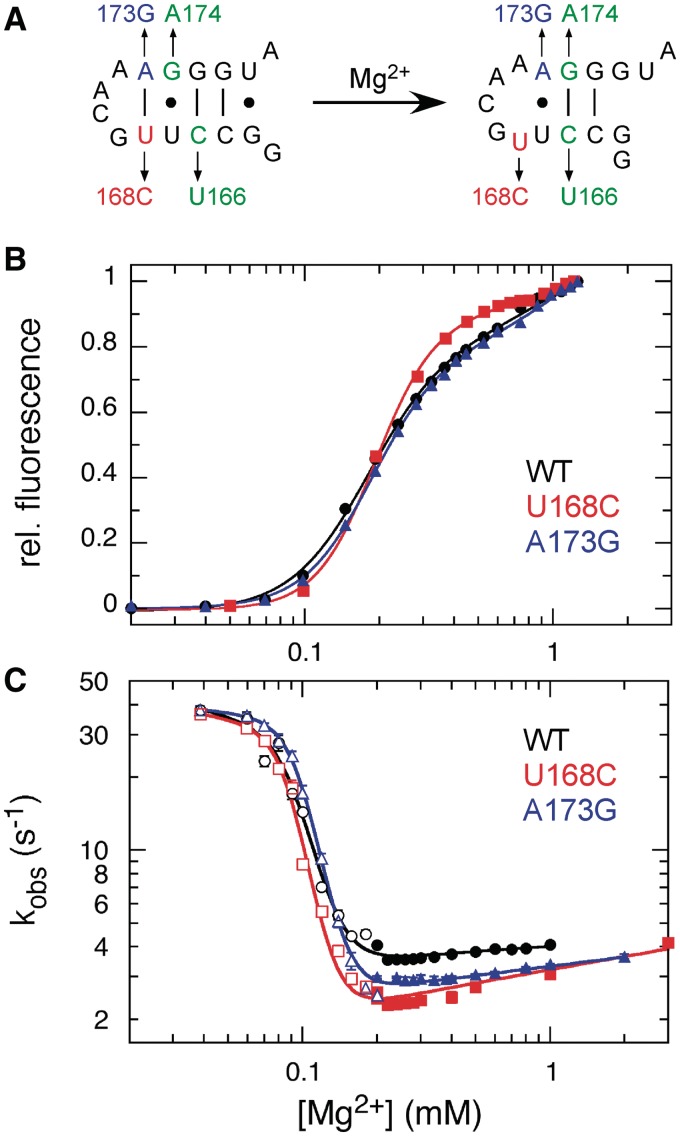
Previous studies showed that mutations which favor the extended conformation of P5c inhibit base pair switching and tertiary folding, while mutations that favor the native secondary structure of P5c stabilize the native state (17). We used mutations that favor the native form of P5c to determine whether P5c switching slows down refolding of tP5abc (Figure 6A). These included U168C, A173G and the double mutation C166U, G174A, all of which disrupt base pairs in the extended P5c helix while maintaining the native base pairs. The mutant RNAs were slightly more stable than the parental RNA (Figure 6B and Supplementary Table S2), confirming a very small shift in their folding equilibria toward the native structure.
Figure 6.
Rearrangement of P5c is not rate-limiting. (A) Base substitutions in P5c predicted to destabilize the extended conformation (left) without destabilizing the native structure (right). (B) Equilibrium Mg2+ titrations of wild-type tP5abc (black, filled circle), U168C (red, filled rhombus), A173G (blue, filled triangle) and G174A, C166U (green). Data were fit to Equation (1), which accounts for changes in cooperativity as well as midpoint (Supplementary Table S2). (C) Folding kinetics in Mg2+ at 30°C. Folding rates were measured as in Figure 4B and fit to Equation (3) (Supplementary Table S3). Open symbols, unfolding; filled symbols, folding conditions.
Although we expected the mutant tP5abc RNAs to fold faster because the native P5c helix is more preferred, they refolded at a similar rate as the parental tP5abc (3 s−1 versus. 4 s−1 in 1 mM Mg2+; Supplementary Table S3). About 15% of the C166U, G174A mutant refolded at a much slower rate (0.64 s−1; Supplementary Figure S6). Like the parental tP5abc, the folding rates did not increase with Mg2+ (Figure 6C). Thus, mutations that favor the native secondary structure of P5c failed to overcome the rate-limiting step of folding. From these results, we concluded that the overall folding rate is determined by reorganization of other interactions within the extended P5abc structure and not the P5c switch. In the subsequent sections, we discuss how the results of these experiments and our simulations can be rationalized by coupled formation of tertiary interactions and helix switching.
DISCUSSION
How secondary and tertiary interactions are coupled in RNA remains an important question, as many RNAs such as riboswitches and even some ribozymes use helix switching to drive regulation or substrate recognition (36,37). When P5abc RNA folds, its secondary structure rearranges to a base pairing scheme that is less energetically favorable but that positions certain bases to participate in tertiary interactions (6). Mg2+ ions stabilize the tertiary interactions both directly and indirectly, compensating for the energetic cost of reorganizing the base pairs and maintaining a net favorable folding free energy (17,18). Thus, remodeling of the RNA secondary structure is also coupled to the specific coordination of metal ions.
Tertiary folding drives helix switching
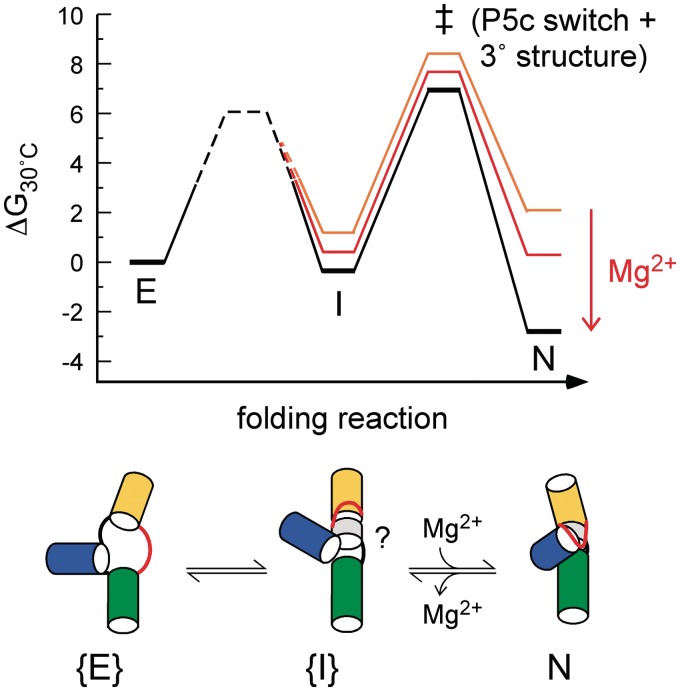
Remarkably, both the simulated equilibrium folding landscapes and the folding kinetics of P5c mutants suggest that P5c switches to its native form just before or concurrently with formation of tertiary interactions in the A-rich bulge (Figure 7). Not only is the native P5c helix predicted to form in the same thermal transition as the tertiary structure (Figure 3) but mutations that destabilize the extended form of P5c (and thus favor the native P5c) failed to speed folding (Figure 6), as would be expected if this switch happens before the tertiary structure forms. The folding kinetics also suggests that P5c switching occurs after the slow step of folding, which itself may involve local tertiary interactions around the three-helix junction (see below). Both aspects of our model are consistent with the proposal that the tertiary interactions directly promote the secondary structure rearrangement (18): the first step creates an intermediate conducive to switching, while the second step couples switching to tertiary interactions between P5c and the A-rich bulge. Ion concentrations, temperature and mutations, which modulate the folding landscapes of P5abc, can be used to control the extent to which tertiary interactions determine P5c switching.
Figure 7.
Model for refolding of tP5abc. The folding pathway from the extended state ensemble {E} to the folded state N involves at least one metastable intermediate ensemble, {I}, which requires partial reorganization of interactions in E and a second step that represents the Mg2+-dependent transition state (‡). The switch in P5c occurs in the second step of the folding reaction, together with the formation of tertiary interactions between P5c and the A-rich bulge. Black, native conditions (1 mM Mg2+); red, midpoint (0.18 mM Mg2+); orange, unfolded conditions (0.05 mM Mg2+). The landscape is based on the equilibrium stability of native tP5abc and simulations of experimental folding kinetics (Berkeley Madonna) assuming an intrinsic reaction time τ0 = 1 µs. The barrier between E and I was poorly constrained (dashed black line).
In our model, the energetically costly P5c switch occurs very late in the folding landscape (Figure 7). As shown in our simulations, the native P5c helix is much less stable than the extended form and thus has little probability of forming in the extended state. Thus, if the P5c switch occurred early, the activation barrier to folding would be very large, and P5abc might never fold. Instead, we suggest earlier secondary and tertiary folding steps couple the change in secondary structure to the formation of tertiary interactions in the A-rich bulge, guiding the RNA through a lower energy path to the native state.
Interestingly, the double mutation G174A, C166U, which stabilizes the native P5c helix relative to the extended form, results in biphasic folding kinetics. This could be due to the presence of an alternative folding path in which P5c switches before the rate-limiting conformational change. Such a possibility is observed in the simulated folding landscapes (Figure 3B).
Folding involves a metastable intermediate
The tP5abc refolding kinetics in Mg2+ were simulated by a two-step folding pathway involving at least one intermediate (I), consistent with high thermal activation energy of Mg2+-induced refolding and the acceleration of folding by urea (Figure 7). Small differences in the observed rate constants of folding and unfolding reactions near the Mg2+ midpoint (Figure 4B) suggest these pathways sample different transition state ensembles consistent with this model.
If P5c switching is not rate-limiting, folding must involve another structural change leading to the I state. This step could involve proper base pairing of P5a, which our simulations show depends on prior formation of the internal P5b and P5c helices in the extended state. The I state may also involve organization of the three-helix junction, including the tandem G-A pairs in P5b which are critical to the native tertiary structure. Substitution of G163 with 2,6-diaminopurine prevented tP5abc RNA from folding (Eda Koculi unpublished data), consistent with its participation in an extensive tertiary hydrogen bonding network. We considered whether the I state and the transition from I to N arises from reorganization of the tertiary interactions after (rather than before) P5c switching. However, this model does not explain why folding is insensitive to Mg2+ concentration and is inconsistent with our simulations and thermal melting results.
An extended transition state
The relative position of the transition state ensemble along the reaction coordinate can be evaluated by comparing the sensitivity of the transition state free energy to Mg2+ (n‡) to that of the overall reaction free energy (nH), which is expressed as β = n‡/nH (38,39). In unfolding conditions, βU = 0.7–1, consistent with a transition state ensemble similar to the unfolded state, while under folding conditions, βF = 0, which is also consistent with a transition state close to the unfolded state (Supplementary Table S3). These observations suggest that the transition states for both folding and unfolding are more open or extended than the native state and do not involve strong, site-specific coordination of metal ions. By contrast, the hairpin ribozyme folds through a compact transition state that is stabilized by Mg2+ similar to the native state (40). Thus P5abc, which must find a low energy path for helix switching, appears to adopt a different folding strategy than RNAs which fold via quasi-parallel docking of two helical domains.
Effect on ribozyme folding
Time-resolved footprinting experiments showed that the P5abc subdomain is the fastest region of the Tetrahymena ribozyme to fold (15). Therefore, the P5abc region likely follows a similar refolding and helix switching pathway in its native context as it does in isolation. However, interactions with other ribozyme sequences modulate formation of tertiary interactions in P5abc, and thus by extension, the rate of P5c switching. First, although the P5abc domain is marginally stable in isolation, extensive tertiary interactions between P5abc and the core of the Tetrahymena ribozyme stabilize the P5abc subdomain in its native context (41). Second, folding of P5abc is retarded in the wild-type ribozyme by premature docking against helices P4 and P6, suggesting that non-specific docking inhibits reorganization of P5c (34). Third, circular permutation of the ribozyme so that P5abc is near the 5′-end of the RNA causes P5abc to fold more rapidly than it does in the wild-type ribozyme (42).
Although P5abc folds more rapidly when it is decoupled from other domains of the ribozyme, the wild-type connections improve folding of the ribozyme core (42,43). Not only does the tertiary structure of P5abc favor the active form of the ribozyme (13,44) but P5c switching is required for the long-range P14 kissing loop interaction between L5c and L2, which stabilizes the ribozyme core (45). As P14 impedes reorganization of the misfolded ribozyme core (46,47), the P5c switch might improve folding overall by delaying formation of P14 (12,48). Our work suggests that the switch kinetics is itself sensitive to alignment of helices in the ribozyme core, perhaps providing feedback on the quality of the fold.
CONCLUSION
In conclusion, our simulations and experiments show that reorganization of the P5abc RNA secondary structure is tightly coupled to the formation of tertiary interactions, which are in turn stabilized by specifically bound Mg2+ ions. Our results strongly suggest that these steps are kinetically linked and possibly concomitant, illustrating how coupled formation of secondary and tertiary structure in RNA can open up new, lower energy pathways to the native structure. Although simultaneous secondary and tertiary folding violates the hierarchy often assumed for RNA folding, it is commonly seen among proteins. Our results further imply that non-native interactions in the extended state aid folding rather than impede it. Because helix switching and P5abc tertiary interactions occur in the same step, this refolding pathway may improve coordination between the stabilizing P14 interaction and proper docking of helices in the ribozyme core. Elucidation of the details of the folding mechanism and the extent of alternative folding pathways will require a combination of ensemble and single molecule experiments and well-designed simulations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–6.
FUNDING
National Institutes of Health (NIH) [R01 GM46686]; National Science Foundation [CHE 09-14033]; a Ruth L. Kirschstein National Research Service Award (to S.S.C.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank M. Deras and A. Werner for initial fluorescence experiments on tP5abc, I. Tinoco for the coordinates of the extended tP5abc, A. Soto and D. Draper for assistance with UV denaturation experiments and D. Barrick for helpful discussions.
REFERENCES
- 1.Thirumalai D, Hyeon C. RNA and protein folding: common themes and variations. Biochemistry. 2005;44:4957–4970. doi: 10.1021/bi047314+. [DOI] [PubMed] [Google Scholar]
- 2.Sosnick TR. Kinetic barriers and the role of topology in protein and RNA folding. Protein Sci. 2008;17:1308–1318. doi: 10.1110/ps.036319.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 4.Adams A, Lindahl T, Fresco JR. Conformational differences between the biologically active and inactive forms of a transfer ribonucleic acid. Proc. Natl Acad. Sci. USA. 1967;57:1684–1691. doi: 10.1073/pnas.57.6.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen AA, Collins RA. Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc. Natl Acad. Sci. USA. 2001;98:7730–7735. doi: 10.1073/pnas.141039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu M, Tinoco I., Jr RNA folding causes secondary structure rearrangement. Proc. Natl Acad. Sci. USA. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gluick TC, Gerstner RB, Draper DE. Effects of Mg2+, K+, and H+ on an equilibrium between alternative conformations of an RNA pseudoknot. J. Mol. Biol. 1997;270:451–463. doi: 10.1006/jmbi.1997.1119. [DOI] [PubMed] [Google Scholar]
- 8.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 9.Butcher SE, Dieckmann T, Feigon J. Solution structure of a GAAA tetraloop receptor RNA. EMBO J. 1997;16:7490–7499. doi: 10.1093/emboj/16.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crothers DM, Cole PE, Hilbers CW, Shulman RG. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J. Mol. Biol. 1974;87:63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- 11.Tinoco IJ, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–261. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 12.Zheng M, Wu M, Tinoco I., Jr Formation of a GNRA tetraloop in P5abc can disrupt an interdomain interaction in the Tetrahymena group I ribozyme. Proc. Natl Acad. Sci. USA. 2001;98:3695–3700. doi: 10.1073/pnas.051608598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Horst G, Christian A, Inoue T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc. Natl Acad. Sci. USA. 1991;88:184–188. doi: 10.1073/pnas.88.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy FL, Cech TR. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J. Mol. Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 15.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 16.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 17.Silverman SK, Zheng M, Wu M, Tinoco I, Jr, Cech TR. Quantifying the energetic interplay of RNA tertiary and secondary structure interactions. RNA. 1999;5:1665–1674. doi: 10.1017/s1355838299991823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thirumalai D. Native secondary structure formation in RNA may be a slave to tertiary folding. Proc. Natl Acad. Sci. USA. 1998;95:11506–11508. doi: 10.1073/pnas.95.20.11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Thirumalai D, Lee N, Woodson SA, Klimov D. Early events in RNA folding. Annu. Rev. Phys. Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 21.Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat. Struct. Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 22.Juneau K, Podell E, Harrington DJ, Cech TR. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA-solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 23.Uchida T, He Q, Ralston CY, Brenowitz M, Chance MR. Linkage of monovalent and divalent ion binding in the folding of the P4-P6 domain of the Tetrahymena ribozyme. Biochemistry. 2002;41:5799–5806. doi: 10.1021/bi020042v. [DOI] [PubMed] [Google Scholar]
- 24.Das R, Travers KJ, Bai Y, Herschlag D. Determining the Mg2+ stoichiometry for folding an RNA metal ion core. J. Am. Chem. Soc. 2005;127:8272–8273. doi: 10.1021/ja051422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liphardt J, Onoa B, Smith SB, Tinoco IJ, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 26.Thirumalai D, Woodson SA. Kinetics of folding of protein and RNA. Acc. Chem. Res. 1996;29:433–439. [Google Scholar]
- 27.Hyeon C, Thirumalai D. Mechanical unfolding of RNA hairpins. Proc. Natl Acad. Sci. USA. 2005;102:6789–6794. doi: 10.1073/pnas.0408314102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honeycutt JD, Thirumalai D. Metastability of the folded states of globular proteins. Proc. Natl Acad. Sci. USA. 1990;87:3526–3529. doi: 10.1073/pnas.87.9.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J. Am. Chem. Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan J, Thirumalai D, Woodson SA. Magnesium-dependent folding of self-splicing RNA: exploring the link between cooperativity, thermodynamics, and kinetics. Proc. Natl Acad. Sci. USA. 1999;96:6149–6154. doi: 10.1073/pnas.96.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pace CN. The stability of globular proteins. CRC Crit. Rev. Biochem. 1975;3:1–43. doi: 10.3109/10409237509102551. [DOI] [PubMed] [Google Scholar]
- 33.Shelton VM, Sosnick TR, Pan T. Applicability of urea in the thermodynamic analysis of secondary and tertiary RNA folding. Biochemistry. 1999;38:16831–16839. doi: 10.1021/bi991699s. [DOI] [PubMed] [Google Scholar]
- 34.Deras ML, Brenowitz M, Ralston CY, Chance MR, Woodson SA. Folding mechanism of the Tetrahymena ribozyme P4-P6 domain. Biochemistry. 2000;39:10975–10985. doi: 10.1021/bi0010118. [DOI] [PubMed] [Google Scholar]
- 35.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 36.Serganov A. The long and the short of riboswitches. Curr. Opin. Struct. Biol. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reymond C, Beaudoin JD, Perreault JP. Modulating RNA structure and catalysis: lessons from small cleaving ribozymes. Cell Mol. Life Sci. 2009;66:3937–3950. doi: 10.1007/s00018-009-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leffler J. Parameters for the description of transition states. Science. 1953;117:340–341. doi: 10.1126/science.117.3039.340. [DOI] [PubMed] [Google Scholar]
- 39.Matouschek A, Fersht AR. Application of physical organic chemistry to engineered mutants of proteins: Hammond postulate behavior in the transition state of protein folding. Proc. Natl Acad. Sci. USA. 1993;90:7814–7818. doi: 10.1073/pnas.90.16.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc. Natl Acad. Sci. USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doherty EA, Herschlag D, Doudna JA. Assembly of an exceptionally stable RNA tertiary interface in a group I ribozyme. Biochemistry. 1999;38:2982–2990. doi: 10.1021/bi982113p. [DOI] [PubMed] [Google Scholar]
- 42.Lease RA, Adilakshmi T, Heilman-Miller S, Woodson SA. Communication between RNA folding domains revealed by folding of circularly permuted ribozymes. J. Mol. Biol. 2007;373:197–210. doi: 10.1016/j.jmb.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heilman-Miller SL, Woodson SA. Perturbed folding kinetics of circularly permuted RNAs with altered topology. J. Mol. Biol. 2003;328:385–394. doi: 10.1016/s0022-2836(03)00304-8. [DOI] [PubMed] [Google Scholar]
- 44.Johnson TH, Tijerina P, Chadee AB, Herschlag D, Russell R. Structural specificity conferred by a group I RNA peripheral element. Proc. Natl Acad. Sci. USA. 2005;102:10176–10181. doi: 10.1073/pnas.0501498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehnert V, Jaeger L, Michel F, Westhof E. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 46.Pan J, Woodson SA. The effect of long-range loop-loop interactions on folding of the Tetrahymena self-splicing RNA. J. Mol. Biol. 1999;294:955–965. doi: 10.1006/jmbi.1999.3298. [DOI] [PubMed] [Google Scholar]
- 47.Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. Exploring the folding landscape of a structured RNA. Proc. Natl Acad. Sci. USA. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Treiber DK, Williamson JR. Concerted kinetic folding of a multidomain ribozyme with a disrupted loop-receptor interaction. J. Mol. Biol. 2001;305:11–21. doi: 10.1006/jmbi.2000.4253. [DOI] [PubMed] [Google Scholar]
- 49.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.