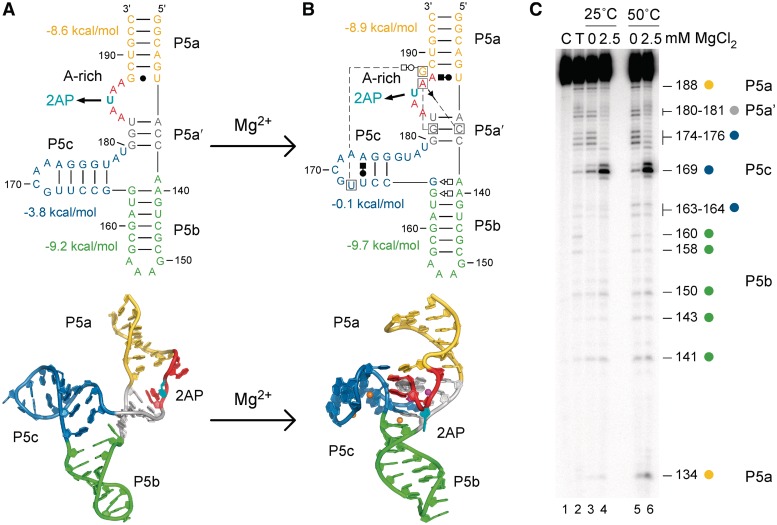
Figure 1.
Structures of the tP5abc RNA. (A) The secondary structures (top) and ribbon diagrams (bottom) of the extended tP5abc from NMR (6) are compared with (B) the folded P5abc structure from X-ray crystallography (22). Mg2+ ions visible in the X-ray structure are represented by spheres. Nucleotides are numbered discontinuously because 5 bp of the natural P5b are missing in tP5abc. Non-Watson–Crick base pairs are represented by Leontis/Westhof symbols (49). U185 was replaced with the fluorescent analog 2AP (cyan), which unstacks in the folded RNA. Base-pairing free energies are from mfold (35). (C) RNase T1 probing of tP5abc secondary structure at 25 and 50°C, with no Mg2+ (extended) or 2.5 mM Mg2+ (folded). Lane C, no digestion control; lane T, RNase T1 sequencing ladder.