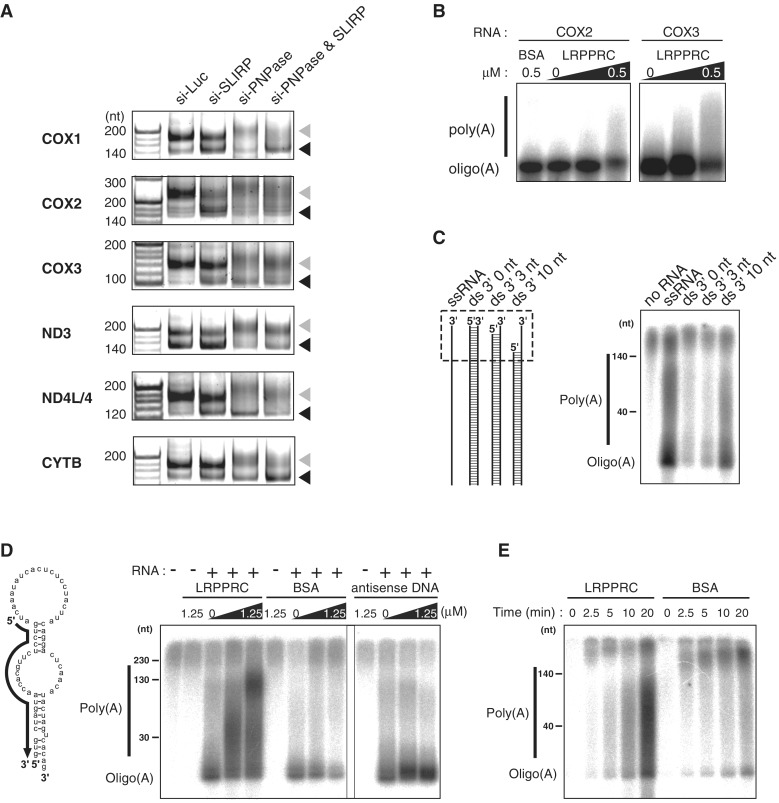
Figure 5.
LRPPRC promotes MTPAP-mediated polyadenylation of mRNAs. (A) Polyadenylation profile of mitochondrial mRNAs upon knockdown of luciferase, SLIRP and PNPase or double knockdown of SLIRP and PNPase. The 3′ termini of the mRNAs for COX1, COX2, COX3, ND3, ND4L/4 and CYTB in HeLa cells transfected with siRNAs were analyzed by the poly(A) tail length assay. RT-PCR products derived from polyadenylated and oligoadenylated RNAs are indicated by gray and black triangles, respectively. The first lanes show 20-nt ladders. (B) In vitro polyadenylation of COX2 or COX3 mRNA transcript catalyzed by recombinant MTPAP in the presence of [α-32P] ATP and different amounts of recombinant LRPPRC (0, 0.1 or 0.5 μM) or BSA (0.5 μM). The products were resolved by polyacrylamide gel electrophoresis and the radioactivity was visualized by using an imaging analyzer (B–E). (C) In vitro polyadenylation of single-stranded (ss) or double-stranded (ds) RNA substrates, catalyzed by recombinant MTPAP in the presence of [α-32P] ATP. Schematic depiction of the substrates is shown on the left. The regions containing 3′ ss overhang of each substrate are boxed in the dotted line. (D) In vitro polyadenylation of the structured RNA (shown on the left) catalyzed by recombinant MTPAP in the presence of [α-32P] ATP and different amounts of recombinant LRPPRC (0, 0.25 or 1.25 μM), BSA (0, 0.25 or 1.25 μM) or antisense DNA (0, 0.25 or 1.25 μM). The complementary region of the antisense oligo, shown as a solid arrow, is depicted on the structured RNA substrate. (E) The time course representation of the in vitro polyadenylation of the ssRNA in the presence of LRPPRC or BSA. 1.25 μM of each protein was added to the reaction mixture.