Figure 9.
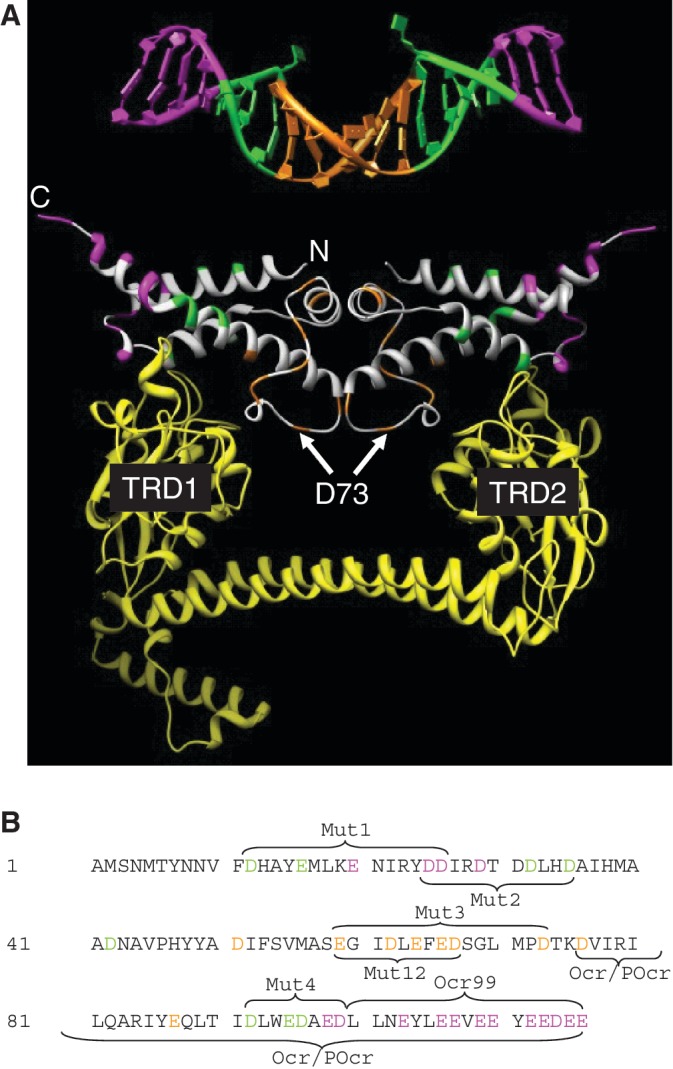
A comparison of the location of the amino acid substitutions made in Ocr with the bent DNA target bound by M.EcoKI (DNA coordinates from 27; pdb 2Y7H). (A) The DNA is coloured magenta for base pairs lying outside of the target sequence, specific base pairs recognized by M.EcoKI (AAC and GTGC) are coloured green and the non-specific spacer of six base pairs is coloured orange. Asp and Glu residues in Ocr equivalent to these regions in DNA are coloured in the same way. Residue D73 found to be important in the Mut3 variant of Ocr is highlighted. The yellow ribbon structure is the S subunit of EcoKI bound to Ocr (27; pdb 2Y7C) showing that the TRDs regions contact the green region in Ocr and the DNA while the alpha helical linkers between the TRDs are close to the orange region of Ocr and the DNA. (B) The sequence of Ocr with the residues equivalent to the magenta, green and orange regions of the DNA in panel A coloured similarly. The regions of the mutations are indicated.
