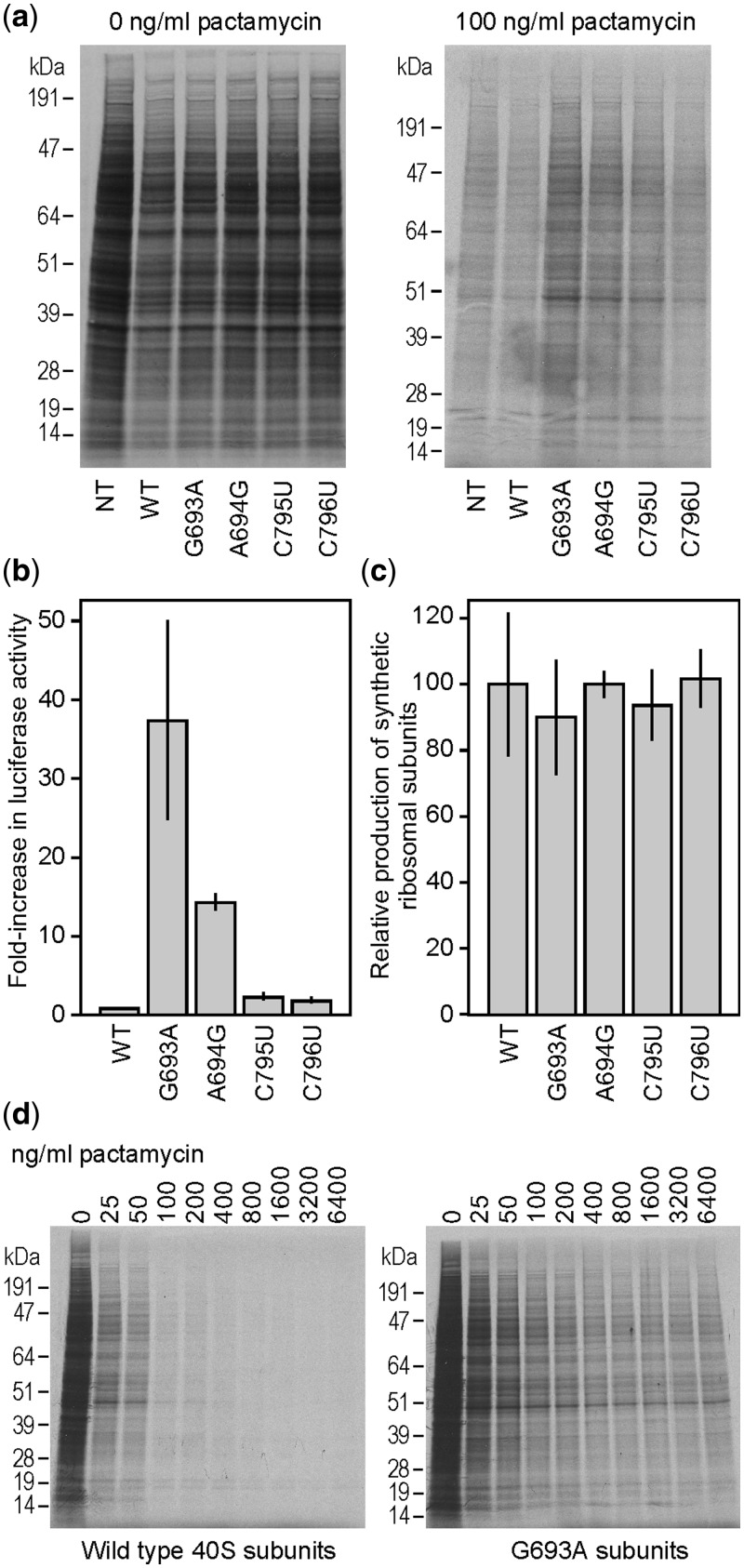
Figure 4.
Analysis of pactamycin-resistance mutations in N2a cells. (a) Protein expression in cells expressing WT or mutated 18 S rRNA constructs. Cells were either not transfected (NT) or transfected with constructs expressing WT 18 S rRNA or 18 S rRNAs containing the point mutations indicated. Cells were then 35S pulse-labeled in the absence or presence of 100 ng/ml pactamycin as indicated, cells were lysed, and equal volumes of the cell lysates were analyzed by SDS-PAGE, as described in Methods. Representative autoradiograms are shown. (b) Relative luciferase activities in cell lysates from cells transfected with WT or mutated 18 S rRNAs and with pGL4.13 (Promega). N2a cells were cotransfected with the 18 S rRNA constructs indicated and with a firefly luciferase construct (pGL4.13) and luciferase expression monitored from cell lysates. Luciferase activities are relative to those obtained from cells transfected with the WT construct, which is set to 1.0. Details of the cotransfection and assay methods are described in Methods. (c) Quantification of synthetic rRNA levels from Northern blots for cells transfected with WT or mutated rRNA constructs. Synthetic rRNA was detected by hybridization with the α-tag probe. Signals were quantified using a Molecular Dynamics Phosphorimager system and are represented relative to WT, with WT set to 100%. (d) Cells were transfected with either WT or G693A mutation-containing 18 S rRNA constructs and incubated with various amounts of pactamycin as indicated. Cells were 35S pulse-labeled, and cell lysates analyzed by SDS-PAGE. Representative autoradiograms are shown. In panels (b) and (c) error bars represent standard deviations from 3 independent experiments.