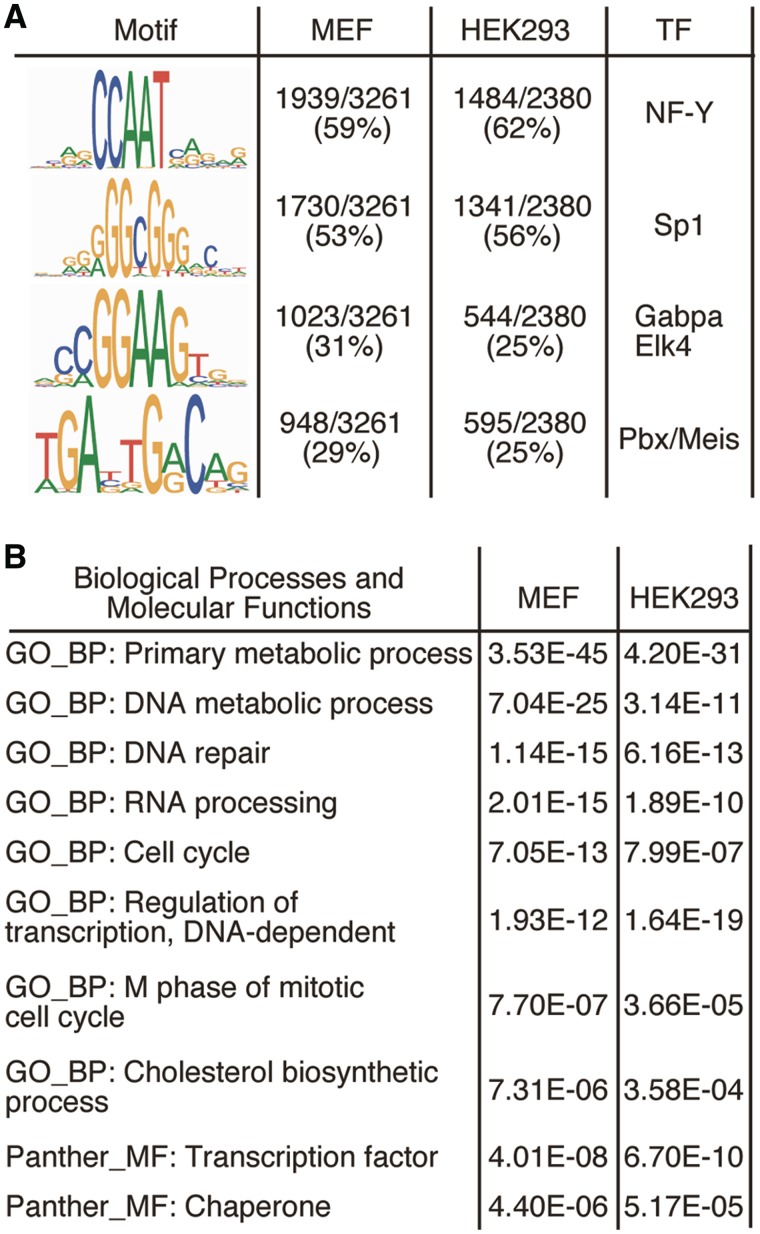
Figure 5.
Computational mining of Sp2-binding sites and target genes. (A) Sequence logos of motifs present in Sp2 ChIP-Seq peaks. Logos were obtained by running MEME (39) with 100 bp summits of all FDR < 0.001 sites. TOMTOM (28) identified these motifs as binding sites for the transcription factors Sp1, NF-Y, Gabpa and Elk4 and Pbx/Meis. (B) Cellular roles of Sp2 target genes. Shown are 10 representative top scoring hits of the pathway analyses along with their Benjamini values.